2. 平凉红牛研究院, 平凉 744000;
3. 平凉市牛产业开发办公室, 平凉 744000
2. Institute of Pingliang Red Cattle, Pingliang 744000, China;
3. Pingliang Cattle Industry Development Office, Pingliang 744000, China
近年来,牛肉作为重要的动物性食品资源,营养价值受到极大的关注,其需求量不断增加,进口量不断增长[1]。在我国严守粮食安全和推进乡村振兴的大背景下,平凉红牛作为优秀的肉牛种质遗传资源,探索其基因表达模式有利于优质地方品种资源的挖掘、保护和利用。牛肉常规营养成分主要包括蛋白质、脂肪和水分等,是影响肉质性状的重要因素,在动物个体间差异很大,反映了品种培育的重要性[2-4]。随着组学技术的发展,转录组测序已成为一种探索复杂性状功能基因的有力工具,关于肉牛肌内脂肪沉积的候选基因已有诸多报道[5-6]。但是,影响肌肉蛋白质和水分含量的候选基因尚不清晰。
本研究以平凉红牛背最长肌为研究对象,测定蛋白质、脂肪和水分的含量,分别挑选表型数据极端差异的高、低组,每组3个重复,通过主成分分析和比较转录组分析,鉴定显著因子载荷基因(significant factor loading genes, SFLGs)和差异表达基因(differentially expressed genes, DEGs)作为候选基因,并进行富集分析。利用所有样本的转录组数据和表型数据进行加权基因共表达网络分析(weighted gene co-expression network analysis, WGCNA),筛选与表型相关的目标模块与Hub基因(hub genes, HGs)。这些结果将为平凉红牛的育种工作提供理论基础。
1 材料与方法 1.1 试验动物及样本采集本试验选用的平凉红牛群体饲养于平凉红牛良种中心,自由采食和饮水,选取40头屠宰月龄(30月龄)及体重(750 kg)相近的阉牛作为试验样本。根据GB/T 19477—2018 《畜禽屠宰操作规程牛》进行屠宰,宰后在左半侧胴体背最长肌的同一位置取样,填入5 mL冻存管后液氮速冻,送至深圳华大基因科技有限公司测序。胴体放置4 ℃冷库排酸72 h后,根据GB/T 27643—2011《牛胴体及鲜肉分割》进行分割,在相同位置取1 kg背最长肌后真空袋封装,转至-20 ℃冷库保存,送至中国农业科学院北京畜牧兽医研究所中心实验室进行营养成分含量测定。
1.2 营养成分测定测定蛋白质、脂肪和水分的含量作为表型数据。蛋白质:参照GB 5009.5—2016《食品中蛋白质的测定》凯氏定氮法。脂肪:参照GB 5009.6—2016《食品中脂肪的测定》酸水解法。水分:参照GB 5009.3—2016《食品中水分的测定》蒸馏法。
1.3 转录组测序使用TRIzol裂解液提取背最长肌的总RNA,通过核酸蛋白测定仪、Nanodrop分光光度计和Agi-lent 2100 bioanalyzer试剂盒检测RNA的浓度、纯度和完整性,RNA浓度大于100 ng·μL-1,RNA纯度大于7.0且OD260 nm/OD280 nm值处于1.8~2.0之间的样品合格,用于测序文库的构建,并在DNBSEQ-T7测序平台进行双端150 bp测序。
通过MD5值检查测序原始数据raw data的完整性,利用Fastp软件质控获得clean data,HISAT2软件将clean data比对到牛的参考基因组,featureCounts和StringTie软件完成定量,获得Counts和FPKM(fragments per kilobase million)[7-10]。
1.4 营养成分含量的相关分析采用R语言进行数据分析,统计结果用最小值、最大值、平均值和标准差表示,通过cor.test函数计算营养成分含量之间的皮尔逊相关系数和显著性。从40头牛分别挑选营养成分含量极端差异的高、低组各3头个体,通过t检验分析高、低组间营养成分含量的差异显著性。
1.5 主成分分析为了探索影响平凉红牛背最长肌营养成分含量的候选基因,高低组共6头个体的FPKM值使用prcomp函数进行主成分分析(PCA),主成分分析计算出第一主成分(PC1)和第二主成分(PC2),随后利用ggplot2将样本绘制在PC1和PC2构建的PCA空间中。如果PCA空间很好地将高、低组区分开,将假设PC1和PC2中超出“μ±5σ”的SFLGs在营养成分含量变化中起重要作用[11]。
1.6 差异表达基因分析使用DESeq2 R包对高、低组Counts进行差异表达分析,筛选标准为FDR < 0.05且log2Fold Change>0.58,以鉴定影响营养成分含量的DEGs[12]。
1.7 富集分析使用clusterProfiler R包对营养成分含量高、低组的SFLGs和DEGs候选基因一起进行GO富集分析[13]。
1.8 加权基因共表达网络分析(WGCNA)WGCNA可用于多样本基因表达量与营养成分含量的关联[14]。通过hclust函数剔除离群样本,pickSoftThreshold函数挑选最佳软阈值,blockwiseModules函数划分基因模块并找到模块特征基因,cor函数和corPvalueStudent函数计算模块特征基因与营养成分含量的皮尔逊相关系数和显著性,皮尔逊相关系数(r)大于0.35且P < 0.05的模块是与营养成分含量相关的目标模块。cor函数计算基因的模块成员度(module membership, MM)和基因重要性(gene significance, GS), 筛选目标模块中MM和GS同时在模块前20%的基因作为HGs。
2 结果 2.1 营养成分含量表型数据的分析本研究中,平凉红牛背最长肌3种常规营养成分含量均在动物个体间有较大差异(表 1)。蛋白质含量最小值为17.23%,最大值为24.48%,平均值为21.42%,标准差为1.51%;脂肪含量最小值为3.50%,最大值为28.19%,平均值为11.47%,标准差为5.21%;水分含量最小值为51.79%,最大值为71.30%,平均值为64.70%,标准差为4.21%。此外,蛋白质、脂肪和水分含量的平均值之和为97.59%,三者涵盖了平凉红牛背最长肌的主要营养成分。
|
|
表 1 平凉红牛背最长肌营养成分含量的测定结果 Table 1 Measurement results of nutrient contents of longissimus dorsi of Pingliang red steer |
3种营养成分含量之间的皮尔逊相关系数如表 2所示,蛋白质含量与水分含量极显著正相关(r=0.60, P < 0.01),蛋白质含量与脂肪含量极显著负相关(r=-0.75, P < 0.01),水分含量与脂肪含量极显著负相关(r=-0.94, P < 0.01)。
|
|
表 2 平凉红牛背最长肌营养成分含量间的皮尔逊相关系数 Table 2 Pearson correlation coefficient between nutrient contents of longissimus dorsi of Pingliang red steer |
如表 3所示,常规营养成分高、低组间的平均含量均显著不同(P < 0.05),表明高、低组的个体选择是合理的。高组3头个体蛋白质含量平均值为23.85%,而低组3头个体蛋白质含量平均值为18.20%,两组间平均含量差异极显著,P=2.26×10-3。高组3头个体脂肪含量平均值为24.23%,而低组3头个体蛋白质含量平均值为4.03%,两组间平均含量差异极显著,P=8.99×10-3。高组3头个体水分含量平均值为70.2%,而低组3头个体蛋白质含量平均值为54.70%,两组间平均含量差异显著,P=1.58×10-2。
|
|
表 3 营养成分含量高、低组的t检验 Table 3 t-test between high and low groups in nutrient contents |
对蛋白质、脂肪和水分含量高、低组个体的FPKM值进行主成分分析,均分别在PC1和PC2构建的二维空间中明显聚类,且PC1和PC2能够解释的方差之和均超过65%,可用于后续的显著因子载荷分析和差异表达分析(图 1)。

|
(A). 蛋白质含量高、低组的主成分分析;(B). 脂肪含量高、低组的主成分分析;(C). 水分含量高、低组的主成分分析。蓝色代表高组,绿色代表低组 (A). Principal component analysis between high and low groups in protein contents; (B). Principal component analysis between high and low groups in fat contents; (C). Principal component analysis between high and low groups in moisture contents. Blue represents the high group and green represents the low group 图 1 营养成分含量高、低组的主成分分析 Fig. 1 Principal component analysis between high and low groups in nutrient contents |
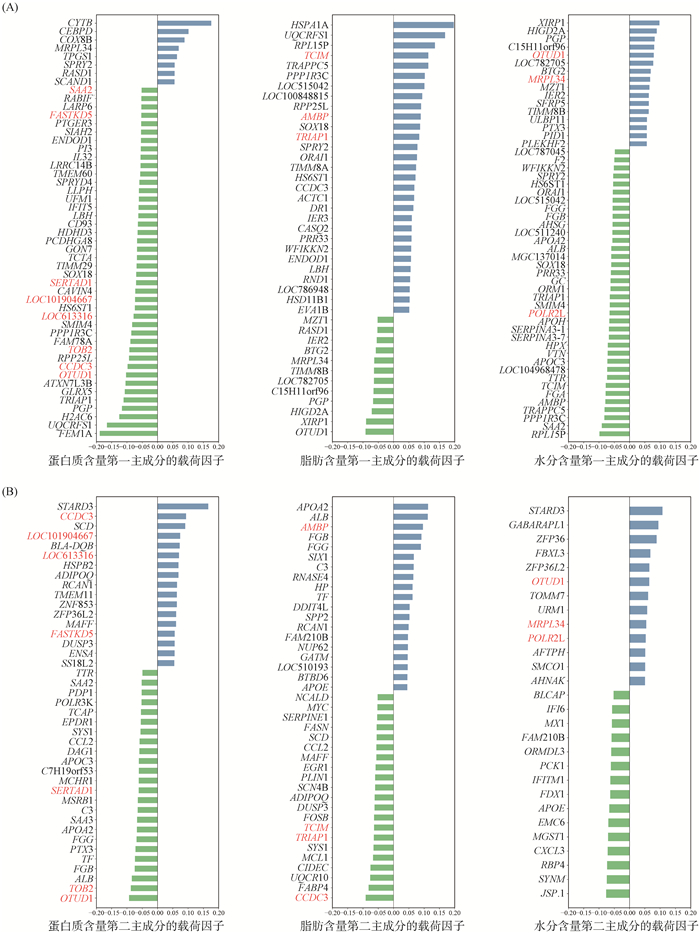
根据大于μ+5σ或小于μ-5σ的标准筛选SFLGs并降序排列,μ为因子载荷的平均值,σ为因子载荷的标准差。PC1中蛋白质含量、脂肪含量和水分含量分别有50、41和52个SFLGs,PC2中分别有41、40和28个SFLGs。筛选PC1和PC2共有的SFLGs,蛋白质中有8个,分别为SAA2、FASTKD5、SERTAD1、LOC101904667、LOC613316、TOB2、CCDC3和OTUD1;脂肪中有4个,分别为TCIM、AMBP、TRIAP1和CCDC3;水分中有3个,分别为OTUD1、MRPL34和POLR2L(图 2)。
|
(A).第一主成分的因子载荷图;(B)第二主成分的因子载荷图。红色字体为前二主成分共有的SFLGs (A).The factor loading diagram of the first principal component; (B).The factor loading diagram of the second principal component. The red font is the SFLGs shared by the first two principal components 图 2 显著因子载荷图 Fig. 2 Significant factor loading diagram |
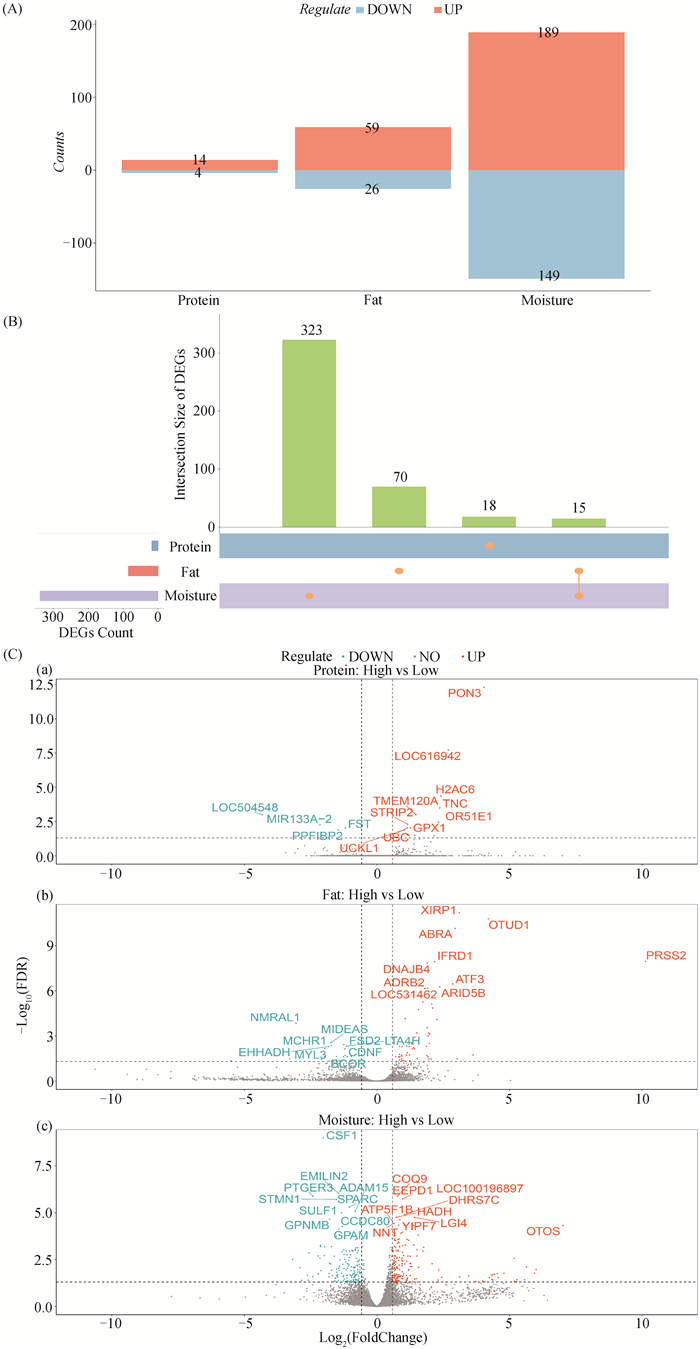
H组相对L组进行差异表达分析,在蛋白质含量高、低组中共有18个差异表达基因,其中14个基因上调,4个基因下调;在脂肪含量高、低组中共有85个差异表达基因,其中59个基因上调,26个基因下调;在水分含量高、低组中共有338个差异表达基因,其中189个基因上调,149个基因下调(图 3A),有15个差异表达基因在脂肪含量和水分含量高、低组中均差异表达(图 3B),3种营养成分中最显著的前10个上调差异表达基因和前10个下调差异表达基因的详细信息在表 4中列出(图 3C)。
|
图 3 蛋白质、脂肪和水分含量高、低组差异表达基因的数量统计图(A)、韦恩图(B)、火山图(C) Fig. 3 The statistical map(A), venn diagram(B), and volcano plot (C) of differentially expressed genes between high and low groups in protein, fat and moisture contents |
|
|
表 4 营养成分含量高、低组的前10个上调和前10个下调差异表达基因 Table 4 The top 10 up-regulated and the top 10 down-regulated differentially expressed genes between high and low groups in nutrient contents |
将营养成分含量高、低组筛选出的SFLGs和DEGs候选基因一起进行GO富集分析。结果显示,蛋白质组只显著富集到一个分子功能:GO: 0031406(羧酸结合);脂肪组全部显著富集:GO: 0050873(棕色脂肪细胞分化)、GO: 0051172(含氮化合物代谢过程的负调控)和GO: 0031324(细胞代谢过程的负调控)等生物过程;水分组富集结果则较为丰富,包括GO: 0022857(跨膜转运蛋白活性)和GO: 0005215(转运蛋白活性)等分子功能,GO: 0071495(细胞对内源性刺激的反应)和GO: 1901655(细胞对酮的反应)等生物过程,GO: 0005581(胶原三聚体)和GO: 0005743(线粒体内膜)等细胞组分(表 5)。
|
|
表 5 营养成分含量高、低组候选基因的富集分析 Table 5 The enrichment analysis of candidate genes between high and low groups in nutrient contents |
为了完善对平凉红牛背最长肌营养成分含量具有重要功能的候选基因的挖掘,筛选显著因子载荷的DEGs,H2AC6为蛋白质含量的候选基因,MAFF、OTUD1和XIRP1为脂肪含量的候选基因,PGP和PID1为水分含量的候选基因(表 6)。
|
|
表 6 营养成分含量高、低组中显著因子载荷的差异表达基因 Table 6 Differentially expressed genes with significant factor loading between high and low groups in nutrient contents |
为了研究营养成分含量与基因表达水平之间的关系,对40头平凉红牛背最长肌的17 383个基因执行加权基因共表达网络分析。本研究中,使用默认参数对40个样本进行聚类,检验离群样本,最终保留所有样本。选择了最优软阈值β=7(图 4A)构建无尺度分布网络(R2=0.90),最小模块的基因数为50,相似度大于0.7的模块合并,最终17 383个基因分配到49个模块中,模块大小从58个基因到3 314个基因(图 4B)。

|
(A).最优软阈值的选择;(B).动态剪切聚类树,不同颜色代表不同模块 (A).The selection of best soft threshold; (B).Dynamic tree cut, different colors represent different modules 图 4 加权基因共表达网络分析 Fig. 4 Weighted gene co-expression network analysis |
为了鉴定到与营养成分含量相关的目标模块,计算每个模块的特征基因与蛋白质含量、脂肪含量和水分含量之间的皮尔逊相关系数和P值,共得到3个皮尔逊相关系数大于0.35且P值小于0.05的目标模块。其中,MEyellowgreen模块为3种营养成分含量共有的目标模块,MEdarkorange2模块为蛋白质含量和脂肪含量共有的目标模块,MElightyellow模块为水分含量独有的目标模块(图 5)。

|
模块基因表达量与营养成分含量关联分析,红色代表正相关,蓝色代表负相关,颜色强度代表相关性程度 The correlation analysis between module gene expression and nutrient contents. Red represents positive correlation, blue represents negative correlation, and color intensity represents correlation degree 图 5 模块性状关联图 Fig. 5 Module-trait relationship diagram |
在目标模块中,蛋白质含量、脂肪含量和水分含量分别筛选到10、14和26个Hub基因,ASIC5、CCDC54、IL36A、MYADML2、OR52E2、TMEM52B和ZBTB14在3种营养成分均存在,其中MYADML2在水分含量高、低组中差异表达(log2Fold Change=0.97,FDR=5.78×10-4),还有3个Hub基因LIN28A、OR6C4和SST在两种营养成分中均存在(表 7)。
|
|
表 7 3种营养成分目标模块的Hub基因 Table 7 Hub genes of the target modules of 3 nutrients |
平凉红牛的生长性能和产肉性能良好,肉质优良,营养成分含量丰富,可为人民提供优质的动物性食品资源,极具开发利用价值[15]。蛋白质、脂肪和水分是牛肉的常规营养成分, 三者之间含量的变化会影响到大理石花纹、嫩度和风味等肉质性状,因此在固定的总量内,这些成分的最佳混合比例可提供最佳的牛肉风味[16]。目前,肉牛相关功能基因的研究主要集中于肌内脂肪沉积,然而肌肉中蛋白质和水分含量相关功能基因的研究仍然较少[17]。因此,本研究以平凉红牛背最长肌为试验对象,探讨影响其蛋白质、脂肪和水分3种营养成分含量的候选基因的表达模式。
蛋白质含量高、低组中,通过主成分分析鉴定到PC1和PC2中共有的8个SFLGs。其中,SAA2是乳蛋白和乳脂性状最有希望的候选基因之一,编码高密度脂蛋白中的载体蛋白组分[18]。FASTKD5位于线粒体RNA颗粒,几乎调控线粒体中所有的mRAN和负责正常的蛋白质合成[19]。真核生物细胞中主要有两条蛋白质降解途径,分别是溶酶体途径和泛素-蛋白酶体途径,在维持肌肉中蛋白质稳态起重要作用[20]。SERTAD1对溶酶体蛋白生物合成具有促进作用,从而对溶酶体的含量和水解酶蛋白质丰度起调节作用[21]。自噬-溶酶体途径的活化与脂肪组织的脂肪分解增加有关,组织蛋白酶B活性会对肌肉蛋白质的质量产生重要影响[22-23]。因此,SERTAD1可能通过溶酶体途径参与平凉红牛背最长肌的蛋白质水解,但这需要进一步研究。此外,OTUD1编码去泛素化酶,OTUD1的上调可通过泛素-蛋白酶体途径加速肌肉组织蛋白质水解[24]。主成分和差异表达进行联合分析,鉴定到蛋白质中1个显著因子载荷的DEGs,H2AC6编码核小体结构的核心组蛋白H2A,与背最长肌可溶性胶原蛋白含量极显著负相关[25]。将蛋白质含量高、低组筛选出的SFLGs和DEGs候选基因一起进行GO富集分析,显著富集到GO: 0031406(羧酸结合),可能与肌少症相关[26]。
脂肪含量高、低组中,通过主成分分析鉴定到PC1和PC2中共有的4个SFLGs。其中,AMBP与内肽酶抑制剂活性相关,从而诱导脂代谢的变化[27]。TRIAP1编码一种线粒体伴侣,与SLMO1的PRELI状结构域结合,构成TRIAP1-SLMO1异二聚体,参与脂质转移[28]。CCDC3表达水平与肌内脂肪含量显著相关,可能参与脂肪或能量代谢[29]。ARID5B是最显著的前10个上调差异表达基因之一,其表达量与脂肪生成正相关[30]。主成分和差异表达进行联合分析,鉴定到蛋白质中3个显著因子载荷的DEGs。XIRP1、OTUD1和MAFF的表达水平与肌内脂肪含量相关,其中XIRP1可能在肌肉生长和分化中发挥关键作用[31-33]。将脂肪含量高、低组筛选出的SFLGs和DEGs候选基因一起进行GO富集分析,ADIPOQ、FABP4、ADRB2和DUSP10显著富集到GO: 0050873(棕色脂肪细胞分化)。
水分含量高、低组中,通过主成分分析鉴定到PC1和PC2中共有的3个SFLGs。其中,POLR2L编码代谢酶,调节脂质、蛋白质和碳水化合物代谢,在保护机体免受热应激损伤中发挥重要作用[34-35]。PTGER3是最显著的前10个下调差异表达基因之一,对肾小管中水的重吸收起抑制作用[36]。主成分和差异表达进行联合分析,鉴定到蛋白质中2个显著因子载荷的DEGs,PID1表达促进脂肪分解,该生化反应会生成水,同时脂肪含量与水分含量极显著负相关(r=-0.94, P < 0.01),揭示了水分含量的候选基因在脂代谢中发挥着潜在作用[37]。将水分含量高、低组筛选出的SFLGs和DEGs候选基因一起进行GO富集分析,富集到GO: 0022857(跨膜转运蛋白活性)和GO: 0005215(转运蛋白活性)等分子功能,其中AQP4编码水通道蛋白-4,形成水专用通道,调节体内水分平衡,AQP9没有富集到生物学条目,但编码的水通道蛋白-9也与水的运输相关(https://www.uniprot.org/);富集到GO: 0005743(线粒体内膜)、GO: 0005740(线粒体被膜)和GO: 0031966(线粒体膜)等细胞组分,其中NDUFA3编码线粒体膜NADH脱氢酶的亚基,该酶在将电子从NADH转移到呼吸链中起作用,而NDUFA4编码细胞色素C氧化酶的亚基,该酶是线粒体电子传递链中的最后一种酶,催化氧气还原为水(https://www.uniprot.org/)。
利用WGCNA鉴定10个影响蛋白质含量、14个影响脂肪含量和26个影响水分含量的Hub基因,ASIC5、CCDC54、IL36A、MYADML2、OR52E2、TMEM52B和ZBTB14在3种营养成分共同被鉴定到。其中,MYADML2参与到精氨酸和脯氨酸代谢、蛋白质消化和吸收以及组氨酸代谢途径,可能在肌肉发育的蛋白质合成与代谢中发挥重要作用,还是脂肪酸C14∶0含量相关的重要候选基因,同时,在水分含量高、低组中差异表达,可能在水分含量变化中发挥着潜在作用[38-39]。ZBTB14编码的蛋白质被鉴定为骨骼肌的转录因子,参与代谢信号的调节[40]。PNLIP是水分含量的Hub基因,编码胰脂肪酶,是一种重要的脂代谢蛋白,可能与PID1相似在脂代谢中发挥着重要作用[41]。
本试验以40头平凉红牛背最长肌为研究对象,测定蛋白质、脂肪和水分3种营养成分含量,结合转录组测序数据,进行了高、低组的主成分和差异表达分析,结合所有样本的WGCNA结果,鉴定了一些有希望影响3种营养成分含量的候选基因,并筛选到几个与多种营养成分含量相关的共有候选基因,加深了对平凉红牛骨骼肌发育分子机制的理解。
4 结论本研究在平凉红牛背最长肌中共鉴定到7个影响蛋白质含量的候选基因SAA2、FASTKD5、SERTAD1、OTUD1、H2AC6、MYADML2和ZBTB14;13个影响脂肪含量的候选基因AMBP、TRIAP1、CCDC3、ARID5B、XIRP1、OTUD1、MAFF、ADIPOQ、FABP4、ADRB2、DUSP10、MYADML2和ZBTB14;10个影响水分含量的候选基因POLR2L、PTGER3、PID1、AQP4、AQP9、NDUFA3、NDUFA4、MYADML2、ZBTB14和PNLIP。其中,OTUD1是蛋白质和脂肪含量共有的候选基因,MYADML2和ZBTB14是与3种营养成分含量均相关的候选基因。这些结果为平凉红牛肉产量的提高和肉品质的提升提供了数据支撑,为中国高档肉牛品种的育种工作提供了理论基础。
| [1] |
GAJAWEERA C, CHUNG K Y, LEE S H, et al. Assessment of carcass and meat quality of longissimus thoracis and semimembranosus muscles of Hanwoo with Korean beef grading standards[J]. Meat Sci, 2020, 160: 107944. DOI:10.1016/j.meatsci.2019.107944 |
| [2] |
KIM N K, CHO S, LEE S H, et al. Proteins in longissimus muscle of Korean native cattle and their relationship to meat quality[J]. Meat Sci, 2008, 80(4): 1068-1073. DOI:10.1016/j.meatsci.2008.04.027 |
| [3] |
MAO Y W, HOPKINS D L, ZHANG Y M, et al. Beef quality with different intramuscular fat content and proteomic analysis using isobaric tag for relative and absolute quantitation of differentially expressed proteins[J]. Meat Sci, 2016, 118: 96-102. DOI:10.1016/j.meatsci.2016.03.028 |
| [4] |
WANG Y J, WANG Z S, HU R, et al. Comparison of carcass characteristics and meat quality between Simmental crossbred cattle, cattle-yaks and Xuanhan yellow cattle[J]. J Sci Food Agric, 2021, 101(9): 3927-3932. DOI:10.1002/jsfa.11032 |
| [5] |
GAO Y H, WANG S Z, MA Y F, et al. Circular RNA regulation of fat deposition and muscle development in cattle[J]. Vet Med Sci, 2022, 8(5): 2104-2113. DOI:10.1002/vms3.857 |
| [6] |
王子渲, 王巧, 张锦, 等. 基于脾脏转录组筛选北京油鸡和广明白鸡抗热应激相关功能基因[J]. 畜牧兽医学报, 2023, 54(5): 1905-1914. WANG Z X, WANG Q, ZHANG J, et al. Transcriptome based screening of functional genes related to heat stress resistance in Beijing you chickens and Guangming broilers[J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(5): 1905-1914. (in Chinese) |
| [7] |
CHEN S F, ZHOU Y Q, CHEN Y R, et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor[J]. Bioinformatics, 2018, 34(17): i884-i890. DOI:10.1093/bioinformatics/bty560 |
| [8] |
KIM D, LANGMEAD B, SALZBERG S L. HISAT: a fast spliced aligner with low memory requirements[J]. Nat Methods, 2015, 12(4): 357-360. DOI:10.1038/nmeth.3317 |
| [9] |
LIAO Y, SMYTH G K, SHI W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features[J]. Bioinformatics, 2014, 30(7): 923-930. DOI:10.1093/bioinformatics/btt656 |
| [10] |
PERTEA M, PERTEA G M, ANTONESCU C M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads[J]. Nat Biotechnol, 2015, 33(3): 290-295. DOI:10.1038/nbt.3122 |
| [11] |
NIHASHI Y, UMEZAWA K, SHINJI S, et al. Distinct cell proliferation, myogenic differentiation, and gene expression in skeletal muscle myoblasts of layer and broiler chickens[J]. Sci Rep, 2019, 9(1): 16527. DOI:10.1038/s41598-019-52946-4 |
| [12] |
LOVE M I, HUBER W, ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15(12): 550. DOI:10.1186/s13059-014-0550-8 |
| [13] |
WU T Z, HU E Q, XU S B, et al. clusterProfiler 4.0:a universal enrichment tool for interpreting omics data[J]. Innovation (Camb), 2021, 2(3): 100141. |
| [14] |
LANGFELDER P, HORVATH S. WGCNA: an R package for weighted correlation network analysis[J]. BMC Bioinformatics, 2008, 9: 559. DOI:10.1186/1471-2105-9-559 |
| [15] |
WANG Z W, AN X J, YANG Y H, et al. Comprehensive analysis of the longissimus dorsi transcriptome and metabolome reveals the regulatory mechanism of different varieties of meat quality[J]. J Agric Food Chem, 2023, 71(2): 1234-1245. DOI:10.1021/acs.jafc.2c07043 |
| [16] |
IIDA F, SAITOU K, KAWAMURA T, et al. Effect of fat content on sensory characteristics of marbled beef from Japanese black steers[J]. Anim Sci J, 2015, 86(7): 707-715. DOI:10.1111/asj.12342 |
| [17] |
DOS SANTOS SILVA D B, FONSECA L F S, PINHEIRO D G, et al. Prediction of hub genes associated with intramuscular fat content in Nelore cattle[J]. BMC Genomics, 2019, 20(1): 520. DOI:10.1186/s12864-019-5904-x |
| [18] |
YANG S, LI C, XIE Y, et al. Detection of functional polymorphisms influencing the promoter activity of the SAA2 gene and their association with milk production traits in Chinese Holstein cows[J]. Anim Genet, 2015, 46(6): 591-598. DOI:10.1111/age.12332 |
| [19] |
ANTONICKA H, SHOUBRIDGE E A. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis[J]. Cell Rep, 2015, 10(6): 920-932. DOI:10.1016/j.celrep.2015.01.030 |
| [20] |
ZIENTARA-RYTTER K, SUBRAMANI S. The roles of ubiquitin-binding protein shuttles in the degradative fate of ubiquitinated proteins in the ubiquitin-proteasome system and autophagy[J]. Cells, 2019, 8(1): 40. DOI:10.3390/cells8010040 |
| [21] |
NGUYEN H A, VU S H, JUNG S, et al. SERTAD1 sensitizes breast cancer cells to doxorubicin and promotes lysosomal protein biosynthesis[J]. Biomedicines, 2022, 10(5): 1148. DOI:10.3390/biomedicines10051148 |
| [22] |
YU H, FAN M H, CHEN X Y, et al. Activated autophagy-lysosomal pathway in dairy cows with hyperketonemia is associated with lipolysis of adipose tissues[J]. J Dairy Sci, 2022, 105(8): 6997-7010. DOI:10.3168/jds.2021-21287 |
| [23] |
LI Z, ZHOU T, WU Y, et al. Investigation of the activity of cathepsin B in red shrimp (Solenocera crassicornis) and its relation to the quality of muscle proteins during chilled and frozen storage[J]. J Food Sci, 2022, 87(4): 1610-1623. DOI:10.1111/1750-3841.16105 |
| [24] |
SKUGOR A, KJOS N P, SUNDARAM A Y M, et al. Effects of long-term feeding of rapeseed meal on skeletal muscle transcriptome, production efficiency and meat quality traits in Norwegian landrace growing-finishing pigs[J]. PLoS One, 2019, 14(8): e0220441. DOI:10.1371/journal.pone.0220441 |
| [25] |
LÓPEZ-PEDROUSO M, LORENZO J M, DI STASIO L, et al. Quantitative proteomic analysis of beef tenderness of piemontese young bulls by SWATH-MS[J]. Food Chem, 2021, 356: 129711. DOI:10.1016/j.foodchem.2021.129711 |
| [26] |
SHAFIEE G, ASGARI Y, SOLTANI A, et al. Identification of candidate genes and proteins in aging skeletal muscle (sarcopenia) using gene expression and structural analysis[J]. PeerJ, 2018, 6: e5239. DOI:10.7717/peerj.5239 |
| [27] |
XU Z Y, CHEN W T, WANG L Y, et al. Cold exposure affects lipid metabolism, fatty acids composition and transcription in pig skeletal muscle[J]. Front Physiol, 2021, 1: 748801. |
| [28] |
MILIARA X, GARNETT J A, TATSUTA T, et al. Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes[J]. EMBO Rep, 2015, 16(7): 824-835. DOI:10.15252/embr.201540229 |
| [29] |
EBERLEIN A, KALBE C, GOLDAMMER T, et al. Analysis of structure and gene expression of bovine CCDC3 gene indicates a function in fat metabolism[J]. Comp Biochem Physiol B Biochem Mol Biol, 2010, 156(1): 19-25. DOI:10.1016/j.cbpb.2010.01.013 |
| [30] |
ZHAO Y H, CHEN S K, YUAN J N, et al. Comprehensive analysis of the lncRNA-miRNA-mRNA regulatory network for intramuscular fat in pigs[J]. Genes (Basel), 2023, 14(1): 168. DOI:10.3390/genes14010168 |
| [31] |
DANG Y L, DONG Q, WU B W, et al. Global landscape of m6A methylation of differently expressed genes in muscle tissue of liaoyu white cattle and simmental cattle[J]. Front Cell Dev Biol, 2022, 10: 840513. DOI:10.3389/fcell.2022.840513 |
| [32] |
ZHANG J, WANG J Y, MA C, et al. Comparative transcriptomic analysis of mRNAs, miRNAs and lncRNAs in the Longissimus dorsi muscles between fat-type and lean-type pigs[J]. Biomolecules, 2022, 12(9): 1294. DOI:10.3390/biom12091294 |
| [33] |
CARDOSO T F, QUINTANILLA R, CASTELLÓ A, et al. Differential expression of mRNA isoforms in the skeletal muscle of pigs with distinct growth and fatness profiles[J]. BMC Genomics, 2018, 19(1): 145. DOI:10.1186/s12864-018-4515-2 |
| [34] |
ÖZMEN Ö, KARAMAN K. Transcriptome analysis and potential mechanisms of bovine oocytes under seasonal heat stress[J]. Anim Biotechnol, 2023, 34(4): 1179-1195. |
| [35] |
ZHAO H H, SOUFAN O, XIA J G, et al. Transcriptome and physiological analysis reveal alterations in muscle metabolisms and immune responses of grass carp (Ctenopharyngodon idellus) cultured at different stocking densities[J]. Aquaculture, 2019, 503: 186-197. DOI:10.1016/j.aquaculture.2019.01.003 |
| [36] |
BRYSON T D, HARDING P. Prostaglandin E2 EP receptors in cardiovascular disease: an update[J]. Biochem Pharmacol, 2022, 195: 114858. DOI:10.1016/j.bcp.2021.114858 |
| [37] |
YIN C Y, LIU W H, LIU Y S, et al. PID1 alters the antilipolytic action of insulin and increases lipolysis via inhibition of AKT/PKA pathway activation[J]. PLoS One, 2019, 14(4): e0214606. DOI:10.1371/journal.pone.0214606 |
| [38] |
HU Z G, LIU X L. Integration of transcriptomics and non-targeted metabolomics reveals the underlying mechanism of skeletal muscle development in duck during embryonic stage[J]. Int J Mol Sci, 2023, 24(6): 5214. DOI:10.3390/ijms24065214 |
| [39] |
PALOMBO V, MILANESI M, SGORLON S, et al. Genome-wide association study of milk fatty acid composition in Italian Simmental and Italian Holstein cows using single nucleotide polymorphism arrays[J]. J Dairy Sci, 2018, 101(12): 11004-11019. DOI:10.3168/jds.2018-14413 |
| [40] |
NYLÉN C, AOI W, ABDELMOEZ A M, et al. IL6 and LIF mRNA expression in skeletal muscle is regulated by AMPK and the transcription factors NFYC, ZBTB14, and SP1[J]. Am J Physiol Endocrinol Metab, 2018, 315(5): E995-E1004. DOI:10.1152/ajpendo.00398.2017 |
| [41] |
CHEN Z T, TENG J Y, DIAO S Q, et al. Insights into the architecture of human-induced polygenic selection in Duroc pigs[J]. J Anim Sci Biotechnol, 2022, 13(1): 99. DOI:10.1186/s40104-022-00751-x |
(编辑 郭云雁)



