2. 江西省幕村农牧科技有限公司, 九江 332438;
3. 北京康普森农业科技有限公司, 北京 102206;
4. 江西农业大学, 南昌 330045
2. Jiangxi Mucun Agriculture and Animal Husbandry Technology Co. Ltd., Jiujiang 332438, China;
3. Beijing Compass Agricultural Technology Co. Ltd., Beijing 102206, China;
4. Jiangxi Agricultural University, Nanchang 330045, China
群体遗传多样性(population genetic diversity)是一个群体内基因组差异的表现,是物种进化的本质。对地方猪进行群体遗传多样性研究,可以了解该品种的起源、进化以及不同品种间的遗传关系,对地方猪的保护及利用具有重要意义。群体的遗传多样性能够使得群体在不断变化的环境中适应和生存下来,一般来说,个体数量越多的群体,遗传多样性越丰富[1-3]。在小群体中,由于个体数量少或者家系少,往往会发生近亲繁殖而导致遗传漂变(genetic drift),减少遗传多样性[4]。对家畜品种的遗传多样性研究有助于个性化制定育种方案,更好的保护群体遗传多样性不丢失。
单核苷酸多态性(single nucleotide polymorphism,SNP)是一种常见的遗传多样性,通过对群体内的个体进行全基因组范围内的SNP比较分析,可以得到该群体的遗传多样性、群体内个体间的亲缘关系以及家系结构等[5-8]。基因芯片,又称cDNA微阵列(microarray)、DNA芯片、寡核苷酸芯片(oligo- chip),属于生物芯片的一种[9]。基因组SNP芯片是一种最早广泛应用的基因组水平遗传标记基因分型方法。SNP芯片由于具有密度大、覆盖广、价格低、数据处理方便等优点应用更为广泛,在鸡[10-13]、牛[14-17]、羊[18-21]、犬[22-23]和马[24]等畜禽方面均有使用。目前使用高密度SNP芯片进行中国地方猪群体遗传多样性的研究较多。在江西地方猪品种研究方面,Wang等[3]通过使用Illumina猪60K SNP芯片扫描,分析发现东乡花猪和玉山黑猪近年来发生过近交的情况,而赣西两头乌猪具有丰富的遗传多样性,且近交个体少;此研究通过构建NJ系统发育树发现,东乡花猪和玉山黑猪都可以划分为9个家系,而赣西两头乌猪可划分为7个家系。在其他中国地方猪品种研究方面,蔡春波等[25]通过高密度SNP芯片研究了马身猪保种群的遗传多样性、群体遗传结构等内容,发现马身猪保种群遗传多样性较丰富,但近交程度高,家系少;黄树文等[26]则发现,两广小耳花猪、大花白猪和蓝塘猪的遗传多样性较低;刘彬等[27]研究了青峪猪保种群体3个世代的遗传结构,发现在闭锁的继代繁育过程中存在群体遗传多样性损失的情况。
中国地方猪品种遗传资源丰富,部分表型优异,但近几十年来,随着国外著名商用猪种及配套系的大量涌入,我国地方猪资源受到巨大的冲击。有的品种已灭绝,一些处于濒危状态的品种群体规模急剧下降。现存的地方品种出现了程度不等的遗传多样性降低和种质退化等现象,我国出现了前所未有的地方猪品种种质资源危机。杭猪是江西省地方猪种,是赣西北花猪的代表性品种,具有性成熟早、发情时间长、繁殖率高、屠宰率高、肉质鲜美等优点[28],但同时存在生长速度慢、瘦肉率低等不足,无法满足市场需求,导致杭猪数量急剧减少,开始出现近亲繁殖现象。针对目前杭猪保种场的保种群家系结构不清,无法进行合理保种等问题,本研究使用“中芯一号”50K SNP芯片对杭猪保种群进行群体遗传结构分析、遗传多样性分析以及家系构建,为杭猪品种资源的保护、选育和利用提供科学依据。
1 材料与方法 1.1 试验动物本研究所用30头杭猪均于2021年6月采集自江西省幕村农牧科技有限公司杭猪保种场,包括4头公猪,26头母猪,所有个体的饲养管理条件一致。采集的耳组织样品存放在装有75%酒精的2 mL冻存管内,并放入超低温冰箱保存备用。
1.2 试验方法1.2.1 耳组织基因组DNA提取与质量检测 本研究使用通用型柱式基因组提取试剂盒(Universal Genomic DNA Kit,康为世纪)进行杭猪耳组织样品的基因组DNA提取。提取步骤主要包括:1)将耳组织剪碎置于1.5 mL离心管中,加入Buffer GTL、Proteinase K后进行水浴消化,向消化完全的样品内加入Buffer GL和无水乙醇后震荡混匀离心,将溶液全部转移到吸附柱内,离心弃废液,向吸附柱中分步加入GW1和GW2并分别离心,弃掉废液后室温彻底晾干,最后加入Buffer GE溶解DNA,离心得到DNA溶液;2)将所得的DNA溶液使用0.8%琼脂糖凝胶进行电泳,检查DNA的完整性;3)使用NanoDrop 2000测定DNA的浓度和质量。
1.2.2 杭猪“中芯一号”SNP分型 将质检合格的杭猪基因组DNA样品委托北京康普森生物技术有限公司使用“中芯一号”芯片进行SNP分型。SNP分型试验流程主要包括:使用NaOH溶液基因组DNA进行碱变性、恒温进行全基因组扩增(37 ℃孵育20~24 h)、DNA片段化、DNA沉淀和重悬、将重悬的DNA片段加到芯片上进行杂交(48 ℃孵育16~24 h)、洗掉非特异性结合的DNA、单碱基延伸染色、数据读取等。
1.2.3 SNP分型结果质量控制 使用Plink(V1.90)软件[29]对得到的SNP分型结果按照下述标准筛除不合格的样本和SNP位点:剔除性染色体上以及未定位染色体的SNPs,剔除检出率(call rate)小于90%的SNPs,剔除最小等位基因频率(MAF)小于0.05的SNPs,剔除哈代-温伯格平衡检验P值小于10-6的标记。
1.2.4 杭猪保种群体的遗传多样性分析 使用Plink(V1.90)软件对质控后的数据进行遗传多样性分析,包括计算杭猪保种群体的MAF、期望杂合度(He)、观察杂合度(Ho)、多态信息含量(PIC),分析杭猪保种群体的遗传多样性。
1.2.5 杭猪保种群体的亲缘关系分析 使用Plink(V1.90)软件中的--homozyg--homozyg-window-snp 50--homozyg-window-het 1--homozyg-window-missing 5 --homozyg-kb 500命令对每个个体的连续性纯合片段(runs of homozygosity,ROH)进行估算。该命令的含义是进行全基因组扫描计算ROH,每个窗口50个SNPs,每个窗口至多一个杂合SNP以及5个错配SNPs,设置最短长度为500 kb。计算每个个体的ROH片段总长度与常染色体基因组总长度的比值得到基于ROH的近交系数(FROH),并使用R软件(https://www.r-project.org/)绘制FROH小提琴图。采用Plink(V1.90)构建状态同源(identity by state,IBS)距离矩阵,采用Gmatrix(V2)软件[30]构建G矩阵,分析保种群体个体间的亲缘关系,并使用R软件绘制IBS距离矩阵和G矩阵结果的热图。
1.2.6 杭猪保种群体的家系结构分析 使用Mega X(V10.2)[31]软件绘制进化树,分析杭猪保种群体中公猪的家系结构。
2 结果 2.1 DNA检测本研究所提取的DNA样品浓度均大于100 ng·μL-1,OD260 nm/OD280 nm为1.8~2.0,表明提取的DNA浓度和纯度均符合要求。经0.8%琼脂糖凝胶电泳检测DNA完整性(图 1),结果显示,DNA条带清晰,无拖尾,表明提取的DNA样品质量较好,符合试验要求。
|
2000M. DNA相对分子质量标准;1~10. 杭猪基因组DNA样品 2000M. The DNA reference marker; 1-10. The genomic DNA sample of Hang pigs 图 1 杭猪基因组DNA琼脂糖凝胶电泳结果 Fig. 1 The agarose gel electrophoresis result of Hang pigs genomic DNA |
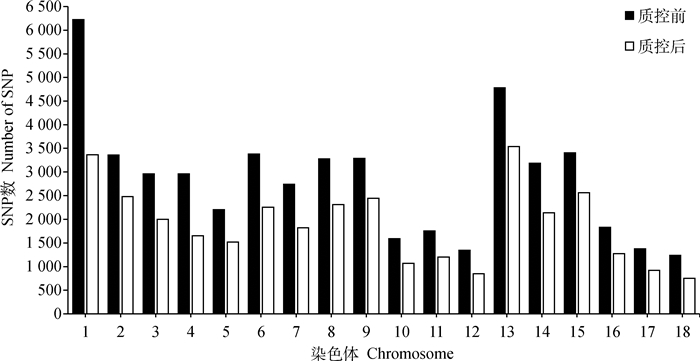
经过“中芯一号”SNP芯片分型,30头杭猪共得到57 466个SNPs标记分型结果,芯片个体检出率为0.987 4~0.989 3,平均检出率为0.988 3± 0.000 4,通过质检的SNPs数为34 156,结果见表 1。质控前后各常染色体上的SNPs数见图 2,其中1号染色体筛选掉的SNP数最多,为2 856个,17号染色体筛选掉的SNP数最少,为454个。
|
|
表 1 SNP质量控制统计结果 Table 1 The statistic results of SNPs after quality control |
|
图 2 质控前后常染色体上SNPs数 Fig. 2 The SNPs number on autosomes before and after quality control |
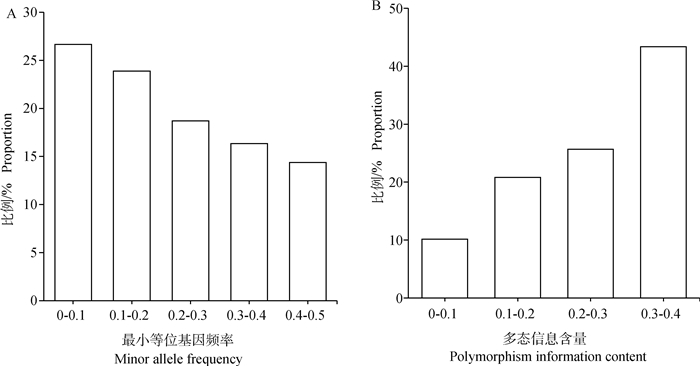
经分析,杭猪保种群体MAF介于0.05到0.1的比例最高,为26.66 %,介于0.4到0.5的比例最低,为14.38%,平均MAF为0.228±0.137,结果见图 3A。SNP位点的多态信息含量(PIC)的范围是0.090~0.375,平均多态信息含量为0.255±0.098,多态信息含量占比见图 3B。杭猪平均观察杂合度(Ho)为0.359±0.181,平均期望杂合度(He)为0.314±0.140,平均观察杂合度略高于平均期望杂合度。
|
A. 最小等位基因频率占比图;B. 多态信息含量占比图 A. The proportion of minor allele frequency; B. The proportion of polymorphism information content 图 3 杭猪保种群体遗传多样性分析结果 Fig. 3 The genetic diversity of Hang pigs conserved population |
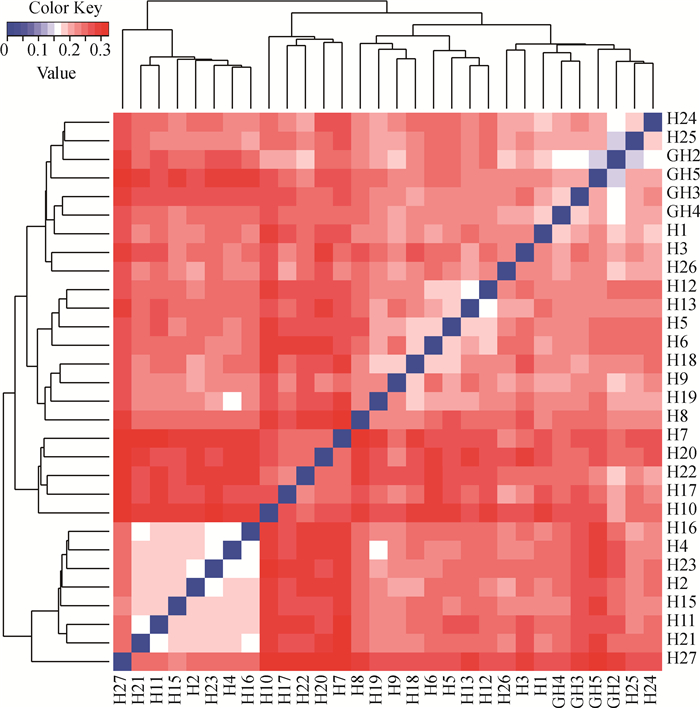
经Plink软件计算,杭猪保种群体的IBS遗传距离在0.143 6~0.323 5之间,平均遗传距离为0.241 7±0.033 6,可视化结果如图 4所示。其中4头公猪(图 4中的GH2、GH3、GH4和GH5)的IBS遗传距离在0.143 6~0.210 9之间,平均遗传距离为0.178 3±0.025 5。从图中可以看出,大部分杭猪个体间的IBS遗传距离较远,亲缘关系处于中等程度(图中偏红的方格);小部分个体间的IBS遗传距离较近,亲缘关系较近(图中偏白以及偏蓝的方格),表明存在近交风险,需强化育种措施。
|
IBS距离矩阵中每一个小方格代表其所连接的两个个体之间的遗传距离值,该值越大越接近红色,即遗传距离越远,反之亦然 Each small square in IBS distance matrix represents the genetic distance value between two connected individuals, the larger the value, the color is closer to red, namely the father the genetic distance is, and vice versa 图 4 杭猪保种群体IBS距离矩阵可视化结果 Fig. 4 The visualization results of IBS distance matrix of Hang pigs conserved population |
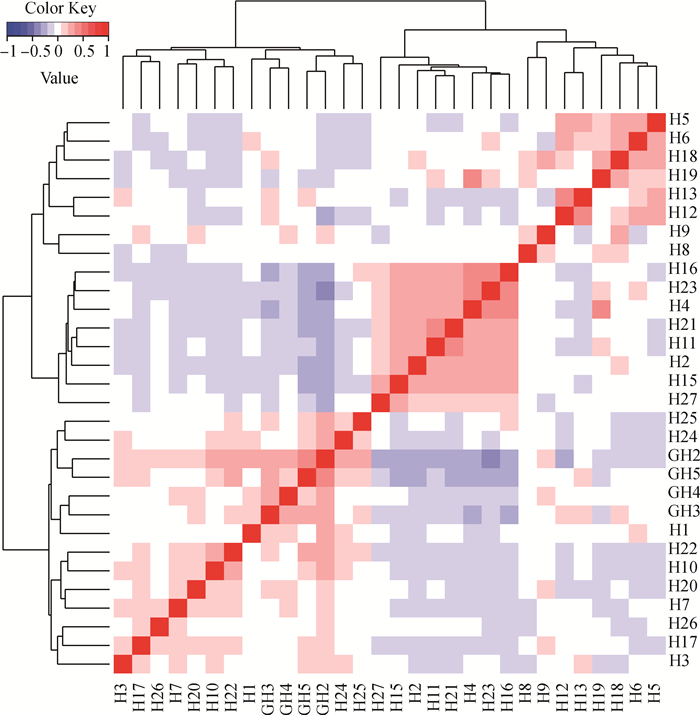
使用Gmatrix软件构建基因组关系G矩阵,进一步分析杭猪保种群体的亲缘关系,可视化结果如图 5所示。从图中可以看到,大部分杭猪个体间亲缘关系程度中等(图中偏白及偏蓝的方格),小部分个体间的亲缘关系较近(图中偏红的方格),与IBS距离矩阵结果一致,表明杭猪保种群体存在近交风险。
|
G矩阵中每一个小方格代表其所连接的两个个体之间的亲缘关系值,该值越大越接近红色,即两个个体的亲缘关系越近,反之亦然 Each small square in G matrix represents the genetic relationship value between two connected individuals, the larger the value, the color is closer to red, namely the closer genetics relationship between two samples, and vice versa 图 5 杭猪保种群体G矩阵可视化结果 Fig. 5 The visualization results of G matrix of Hang pigs conserved population |
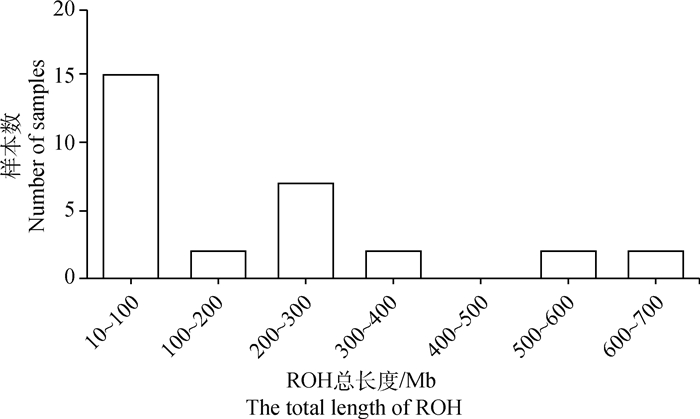
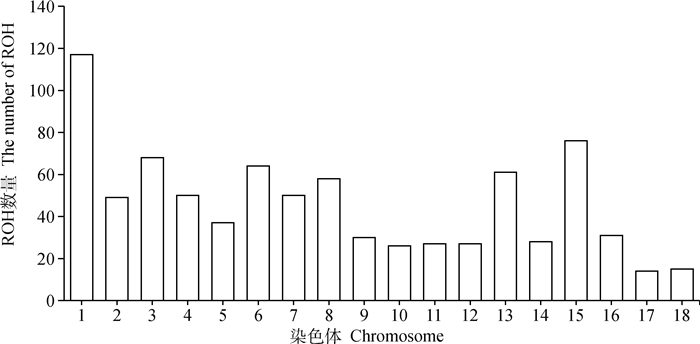
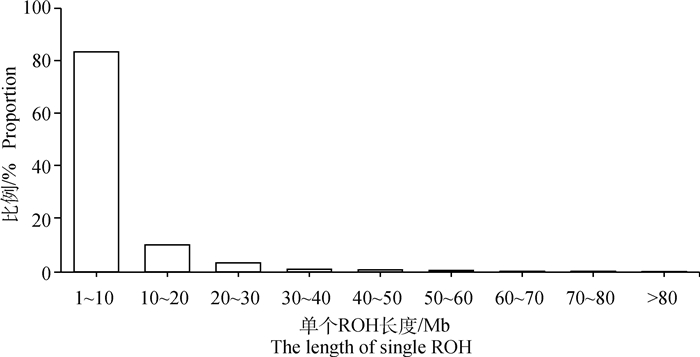
经Plink软件计算,30头杭猪保种个体共检测到828个ROHs,单个ROH平均长度为(6.96±9.62)Mb,个体ROH数量为5~ 80个不等,平均(27.60±19.64)个。个体ROH总长度为(14.14~687.31)Mb,平均(191.95±201.56)Mb,个体间的差异较大,其中ROH总长度在100 Mb以内的个体数量最多,占比50%,而ROH总长度为400~500 Mb的个体数量为0(图 6)。从染色体上的ROH分布数量看,1号染色体上的ROH数量最多,为117个;17号染色体上的ROH数量最少,为14个(图 7)。从单个ROH长度数量分布情况来看,1~10 Mb长度的ROH数量最多,占总数量的83.33%,而大于80 Mb长度的ROH数量最少,只占总数量的0.12%(图 8)。通过对每个个体的ROH统计,得到每个个体基于ROH的近交系数FROH(图 9),计算结果表明,杭猪保种群体的平均FROH为0.078±0.082,但4头公猪中有3头公猪的FROH大于0.21,另1头则大于0.10,公猪平均近交系数为0.219±0.082。由图 9可知,杭猪保种群体整体的近交系数较小,说明近交程度不严重。
|
图 6 杭猪ROH总长度样本数分布 Fig. 6 The distribution of ROH total length in Hang pigs |
|
图 7 杭猪染色体ROH数量分布图 Fig. 7 The distribution of ROH on chromosomes of Hang pigs |
|
图 8 杭猪ROH长度占比统计 Fig. 8 The statistics on proportion of ROH length in Hang pigs |
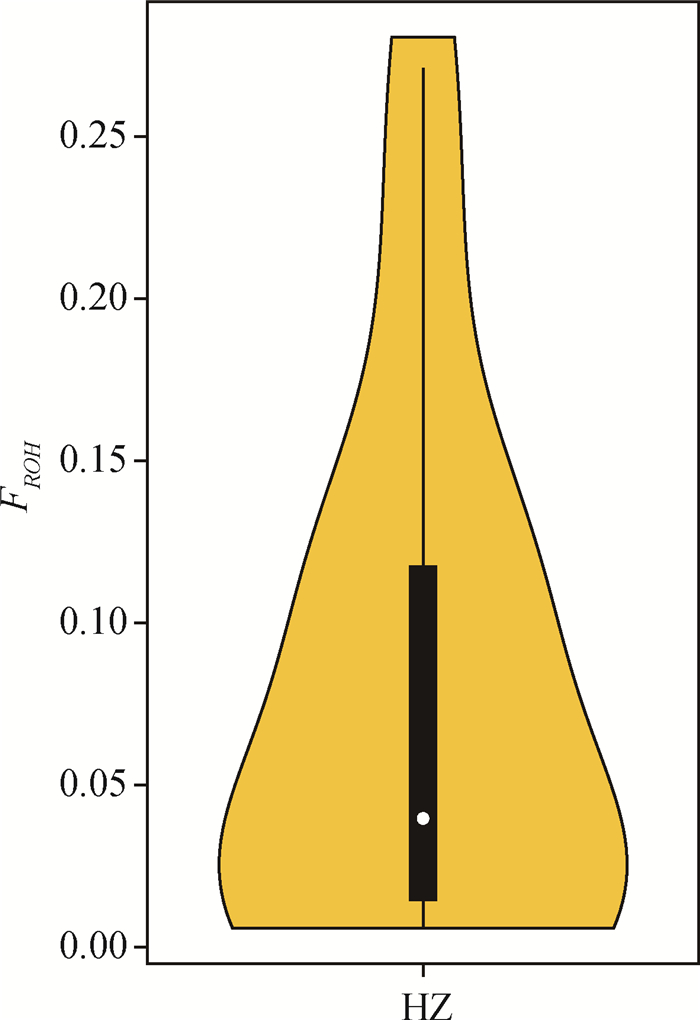
|
图中中心的白点代表杭猪群体FROH的中位数,中间黑色方框的上缘和下缘分别为杭猪群体FROH的上四分位数和下四分位数。小提琴图的宽窄表示群体FROH的样本数量分布,小提琴图越宽的部分表示处于该值的样本数目越多,反之则越少 The white dot in the center of the figure indicates the median of FROH in Hang pigs, and the upper and lower edges of the black box in the middle are the upper quartile and lower quartile of FROH in Hang pigs, respectively. The width of the violin graph represents the sample number of FROH in Hang pigs, the wider indicate the more samples at this FROH value, and vice versa 图 9 基于ROH的近交系数可视化结果 Fig. 9 The visualization result of inbreeding coefficient based on ROH |
对保种群体中的公猪进行家系分类,有助于杭猪的保种和育种工作。结合IBS距离矩阵和G矩阵对4头杭猪公猪使用邻接法进行了聚类分析,使用Mega X软件绘制进化树,结果发现4头公猪都属于同一家系A(图 10)。通过G矩阵结果,将与公猪的亲缘关系值大于0.1的母猪与公猪划分为同一个家系,分析结果表明,家系A中共有4头公猪,12头母猪,其余14头母猪则划分为其它家系,杭猪保种群体家系划分结果见表 2。
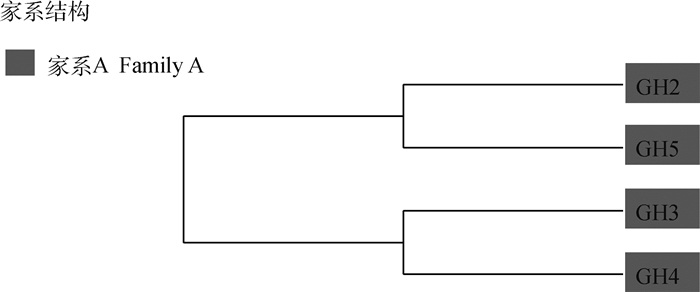
|
图中数字表示公猪号,相同颜色表示为同一个家系 The numbers in the figure indicate the number of boar, and the same color represent the same family 图 10 杭猪保种群体公猪进化树结果 Fig. 10 The phylogenetic tree of boars in Hang pigs conserved population |
|
|
表 2 杭猪保种群体家系表 Table 2 The genealogy of Hang pigs conserved population |
中国地方猪拥有比现代西方猪更长远的家养历史[32],而且历史上中国地方猪没有像西方猪那样进行高强度的人工选择,因此有理由推测中国地方猪的遗传多样性比西方猪丰富,这一观点也得到了前人研究的验证[33-36]。然而本研究结果表明,杭猪的遗传多样性与西方猪的接近[37-38],可能的原因是:1)不同研究使用的芯片不一致,一个是偏向中国地方猪设计的50K SNP芯片,另一个是针对西方猪设计的60K或者70K SNP芯片;2)各研究所使用的个体数不一致,不能完全代表各品种的遗传多样性;3)对SNP芯片分型结果所使用的质控条件不一致;4)据本研究结果可知,杭猪家系少,且部分个体近交系数较高,这也会导致群体遗传多样性降低。
观察杂合度(Ho)为某个品种内所有SNPs位点观察到的杂合基因型个体数占总体数的比例,期望杂合度(He)为某个品种所有SNPs位点杂合基因型的理论比例。当观察杂合度低于期望杂合度时,表明该群体可能存在近交;观察杂合度高于期望杂合度时,表明该群体可能渗入了其它品种的血缘。本研究中,杭猪Ho为0.359,高于东乡花猪(0.17)、玉山黑猪(0.22)、赣西两头乌猪(0.24)、陆川猪(0.18)、巴马香猪(0.22)等的Ho[3],与马身猪(0.354)[25]和凉山猪(0.355)[5]等的Ho接近。杭猪He为0.314,低于梅山猪(0.382)、二花脸(0.378)、米猪(0.382)、嘉兴黑猪(0.350)、沙乌头猪(0.371)[39]、马身猪(0.350)[25]和凉山猪(0.348)[5]的He,与枫泾猪(0.315)[39]的He接近。杭猪的Ho略高于He,说明当前的杭猪群体可能混入过其他品种的血缘,需要加以纯化。
IBS是指两个或两个以上的个体具有相同的等位基因序列,IBS遗传距离可以衡量群体内个体间的相似性,评价彼此的亲缘关系。青峪猪以及马身猪的公猪平均IBS遗传距离均高于保种群体的平均IBS遗传距离[25, 27],而本研究杭猪公猪的平均IBS遗传距离小于杭猪保种群体的平均IBS遗传距离,是因为青峪猪和马身猪公猪分别有6个和3个家系,而本研究的杭猪公猪只有一个家系,因此IBS遗传距离较近。
ROH是指父母将相同的单倍型传递给后代导致该后代中存在的纯合基因型的连续片段。近亲繁殖将导致ROH的增加,现代基因组数据研究显示ROH在群体中普遍存在,群体的ROH的长度和数量反映了群体的人口历史和血统情况。ROH长度过大将导致群体遗传多样性的降低,对群体ROH的估算能更好的确定近亲繁殖情况,为种群多样性保护提供重要参考依据[40]。本研究中,个体ROH总长大于500 Mb的个体只有4个,其中包含3头公猪,这3头公猪基于ROH的近交系数都大于0.21,另一头公猪的ROH总长度为257 Mb,基于ROH的近交系数为0.10,表明这4头公猪存在近交,这与4头公猪为一个家系的结果相符。26头杭猪母猪单个ROH长度小于10 Mb的比例占母猪整体单个ROH数量的86.63%,且母猪的近交系数也偏小,说明杭猪母猪群目前近交程度较轻,需继续加以维持,同时需预防近交的发生。
公猪在群体保种过程中起着十分重要的作用,家系的划分都是以公猪为主,若某个群体的公猪只有一个家系,随着世代的增加和闭锁繁育,近交系数会越来越大[27],种群多样性会降低,健康和繁殖性能下降,隐形有害突变会增加,导致整个种群出现衰退现象,严重的可能导致该物种的灭绝[41-43]。扩增群体数量特别是引入新的公猪血缘是有效降低杭猪近交系数的方法之一,引种后还需加强管理,制定合理的配种方案,做好详细的系谱记录。如果引种困难,可利用与杭猪公猪不属于同一家系的其他家系的母猪通过回交法创建新的公猪血缘,也可以有效避免近交系数的进一步增加。
4 结论本研究通过使用高密度SNP芯片较为系统的分析了杭猪保种场中保种群的遗传多样性、个体间的遗传距离和亲缘关系以及群体的家系结构和近交系数。表明该保种场的杭猪群体多态性处于中等水平;杭猪保种群可能混入过其他品种的血缘,需要进行血缘纯度的鉴定;杭猪公猪遗传距离短,亲缘关系较近,且存在近交,家系少,需要引入新的血统来扩增杭猪保种群血缘数量或者通过回交来降低近交系数,确保杭猪保种群家系结构的合理与平衡。
| [1] |
QIAO R, LI X, HAN X, et al. Population structure and genetic diversity of four Henan pig populations[J]. Anim Genet, 2019, 50(3): 262-265. DOI:10.1111/age.12775 |
| [2] |
VAN BA N, NAM L Q, DO D N, et al. An assessment of genetic diversity and population structures of fifteen Vietnamese indigenous pig breeds for supporting the decision making on conservation strategies[J]. Trop Anim Health Prod, 2020, 52(3): 1033-1041. DOI:10.1007/s11250-019-02090-y |
| [3] |
WANG X, WANG C, HUANG M, et al. Genetic diversity, population structure and phylogenetic relationships of three indigenous pig breeds from Jiangxi Province, China, in a worldwide panel of pigs[J]. Anim Genet, 2018, 49(4): 275-283. DOI:10.1111/age.12687 |
| [4] |
BLACKBURN H D, PLANTE Y, ROHRER G, et al. Impact of genetic drift on access and benefit sharing under the Nagoya protocol: the case of the Meishan pig[J]. J Anim Sci, 2014, 92(4): 1405-1411. DOI:10.2527/jas.2013-7274 |
| [5] |
LIU B, SHEN L Y, GUO Z X, et al. Single nucleotide polymorphism-based analysis of the genetic structure of Liangshan pig population[J]. Anim Biosci, 2021, 34(7): 1105-1115. DOI:10.5713/ajas.19.0884 |
| [6] |
XU P, WANG X P, NI L G, et al. Genome-wide genotyping uncovers genetic diversity, phylogeny, signatures of selection, and population structure of Chinese Jiangquhai pigs in a global perspective[J]. J Anim Sci, 2019, 97(4): 1491-1500. DOI:10.1093/jas/skz028 |
| [7] |
JOAQUIM L B, CHUD T C S, MARCHESI J A P, et al. Genomic structure of a crossbred Landrace pig population[J]. PLoS One, 2019, 14(2): e0212266. DOI:10.1371/journal.pone.0212266 |
| [8] |
ZHOU M L, WANG G F, CHEN M H, et al. Genetic diversity and population structure of sheep (Ovisaries) in Sichuan, China[J]. PLoS One, 2021, 16(9): e0257974. DOI:10.1371/journal.pone.0257974 |
| [9] |
王淑辉. 利用基因芯片对阉割引起牛生长发育和脂肪沉积变化分子机理的研究[D]. 北京: 中国农业科学院, 2008. WANG S H. The study on the molecular mechanism of growth and fatty deposition by genechip in beef cattle[D]. Beijing: Chinese Academy of Agricultural Sciences, 2008. (in Chinese) |
| [10] |
STRILLACCI M G, VEGA-MURILLO V E, ROMÁN-PONCE S I, et al. Looking at genetic structure and selection signatures of the Mexican chicken population using single nucleotide polymorphism markers[J]. Poult Sci, 2018, 97(3): 791-802. DOI:10.3382/ps/pex374 |
| [11] |
NIE C S, ALMEIDA P, JIA Y X, et al. Genome-wide single-nucleotide polymorphism data unveil admixture of Chinese indigenous chicken breeds with commercial breeds[J]. Genome Biol Evol, 2019, 11(7): 1847-1856. DOI:10.1093/gbe/evz128 |
| [12] |
CENDRON F, MASTRANGELO S, TOLONE M, et al. Genome-wide analysis reveals the patterns of genetic diversity and population structure of 8 Italian local chicken breeds[J]. Poult Sci, 2021, 100(2): 441-451. DOI:10.1016/j.psj.2020.10.023 |
| [13] |
STRILLACCI M G, COZZI M C, GORLA E, et al. Genomic and genetic variability of six chicken populations using single nucleotide polymorphism and copy number variants as markers[J]. Animal, 2017, 11(5): 737-745. DOI:10.1017/S1751731116002135 |
| [14] |
BOUSHABA N, BOUJENANE I, MOAZAMI-GOUDARZI K, et al. Genetic diversity and relationships among six local cattle populations in semi-arid areas assessed by a bovine medium-density single nucleotide polymorphism data[J]. Animal, 2019, 13(1): 8-14. DOI:10.1017/S1751731118001179 |
| [15] |
SUEZAWA R, NIKADORI H, SASAKI S. Genetic diversity and genomic inbreeding in Japanese black cows in the islands of Okinawa Prefecture evaluated using single-nucleotide polymorphism array[J]. Anim Sci J, 2021, 92(1): e13525. |
| [16] |
KIM S, CHEONG H S, SHIN H D, et al. Genetic diversity and divergence among Korean cattle breeds assessed using a BovineHD single-nucleotide polymorphism chip[J]. Asian-Australas J Anim Sci, 2018, 31(11): 1691-1699. DOI:10.5713/ajas.17.0419 |
| [17] |
KING F J M, BANGA C B, VISSER C. Genetic diversity and population structure of three native cattle populations in Mozambique[J]. Trop Anim Health Prod, 2021, 53(1): 117. DOI:10.1007/s11250-021-02562-0 |
| [18] |
MEYERMANS R, GORSSEN W, WIJNROCX K, et al. Unraveling the genetic diversity of Belgian milk sheep using medium-density SNP genotypes[J]. Anim Genet, 2020, 51(2): 258-265. DOI:10.1111/age.12891 |
| [19] |
GRASSO A N, GOLDBERG V, NAVAJAS E A, et al. Genomic variation and population structure detected by single nucleotide polymorphism arrays in Corriedale, Merino and Creole sheep[J]. Genet Mol Biol, 2014, 37(2): 389-395. DOI:10.1590/S1415-47572014000300011 |
| [20] |
BERIHULAY H, LI Y, LIU X, et al. Genetic diversity and population structure in multiple Chinese goat populations using a SNP panel[J]. Anim Genet, 2019, 50(3): 242-249. DOI:10.1111/age.12776 |
| [21] |
MOOSANEZHAD KHABISI M, ASADI FOOZI M, LV F H, et al. Genome-wide DNA arrays profiling unravels the genetic structure of Iranian sheep and pattern of admixture with worldwide coarse-wool sheep breeds[J]. Genomics, 2021, 113(6): 3501-3511. DOI:10.1016/j.ygeno.2021.07.019 |
| [22] |
JANEŠ M, ZORC M, CUBRIC-CURIK V, et al. Population structure and genetic history of Tibetan Terriers[J]. Genet Sel Evol, 2019, 51(1): 79. DOI:10.1186/s12711-019-0520-4 |
| [23] |
LETKO A, MINOR K M, JAGANNATHAN V, et al. Genomic diversity and population structure of the Leonberger dog breed[J]. Genet Sel Evol, 2020, 52(1): 61. DOI:10.1186/s12711-020-00581-3 |
| [24] |
COSGROVE E J, SADEGHI R, SCHLAMP F, et al. Genome diversity and the origin of the Arabian horse[J]. Sci Rep, 2020, 10(1): 9702. DOI:10.1038/s41598-020-66232-1 |
| [25] |
蔡春波, 张雪莲, 张万峰, 等. 运用SNP芯片评估马身猪保种群体的遗传结构[J]. 畜牧兽医学报, 2021, 52(4): 920-931. CAI C B, ZHANG X L, ZHANG W F, et al. Evaluation of genetic structure in Mashen pigs conserved population based on SNP chip[J]. Acta Veterinaria et Zootechnica Sinica, 2021, 52(4): 920-931. (in Chinese) |
| [26] |
黄树文, 张哲, 陈赞谋, 等. 广东省现有5个地方猪种基于SNP芯片的遗传多样性分析[J]. 中国畜牧杂志, 2018, 54(6): 33-37. HUANG S W, ZHANG Z, CHEN Z M, et al. Geneticdiversity analysis of five Cantonese indigenous pigs based on SNP chip[J]. Chinese Journal of Animal Science, 2018, 54(6): 33-37. (in Chinese) |
| [27] |
刘彬, 沈林園, 陈映, 等. 基于SNP芯片分析青峪猪保种群体的遗传结构[J]. 畜牧兽医学报, 2020, 51(2): 260-269. LIU B, SHEN L Y, CHEN Y, et al. Analysis of genetic structure of conservation population in Qingyu pig based on SNP chip[J]. Acta Veterinaria et Zootechnica Sinica, 2020, 51(2): 260-269. (in Chinese) |
| [28] |
李文. 不同饲喂模式对杭猪生产性能影响的研究[D]. 南昌: 江西农业大学, 2016. LI W. The study on production performance of different raise model to Xiushui Hang-pig[D]. Nanchang: Jiangxi Agricultural University, 2016. (in Chinese) |
| [29] |
CHANG C C, CHOW C C, TELLIER L C, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets[J]. Giga Science, 2015, 4: 7. DOI:10.1186/s13742-015-0047-8 |
| [30] |
SU G, CHRISTENSEN O F, JANSS L, et al. Comparison of genomic predictions using genomic relationship matrices built with different weighting factors to account for locus-specific variances[J]. J Dairy Sci, 2014, 97(10): 6547-6559. DOI:10.3168/jds.2014-8210 |
| [31] |
KUMAR S, STECHER G, LI M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Mol Biol Evol, 2018, 35(6): 1547-1549. DOI:10.1093/molbev/msy096 |
| [32] |
LARSON G, LIU R R, ZHAO X B, et al. Patterns of East Asian pig domestication, migration, and turnover revealed by modern and ancient DNA[J]. Proc Natl Acad Sci U S A, 2010, 107(17): 7686-7691. DOI:10.1073/pnas.0912264107 |
| [33] |
FAN B, WANG Z G, LI Y J, et al. Genetic variation analysis within and among Chinese indigenous swine populations using microsatellite markers[J]. Anim Genet, 2002, 33(6): 422-427. DOI:10.1046/j.1365-2052.2002.00898.x |
| [34] |
SANCRISTOBAL M, CHEVALET C, HALEY C S, et al. Genetic diversity within and between European pig breeds using microsatellite markers[J]. Anim Genet, 2006, 37(3): 189-198. DOI:10.1111/j.1365-2052.2005.01385.x |
| [35] |
LAVAL G, IANNUCCELLI N, LEGAULT C, et al. Genetic diversity of eleven European pig breeds[J]. Genet Sel Evol, 2000, 32(2): 187-203. DOI:10.1186/1297-9686-32-2-187 |
| [36] |
YANG S L, WANG Z G, LIU B, et al. Genetic variation and relationships of eighteen Chinese indigenous pig breeds[J]. Genet Sel Evol, 2003, 35(6): 657-671. DOI:10.1186/1297-9686-35-7-657 |
| [37] |
AI H S, HUANG L S, REN J. Genetic diversity, linkage disequilibrium and selection signatures in Chinese and western pigs revealed by genome-wide SNP markers[J]. PLoS One, 2013, 8(2): e56001. DOI:10.1371/journal.pone.0056001 |
| [38] |
MUÑOZ M, BOZZI R, GARCÍA-CASCO J, et al. Genomic diversity, linkage disequilibrium and selection signatures in European local pig breeds assessed with a high density SNP chip[J]. Sci Rep, 2019, 9(1): 13546. DOI:10.1038/s41598-019-49830-6 |
| [39] |
WANG Z, CHEN Q, YANG Y, et al. Genetic diversity and population structure of six Chinese indigenous pig breeds in the Taihu Lake region revealed by sequencing data[J]. Anim Genet, 2015, 46(6): 697-701. DOI:10.1111/age.12349 |
| [40] |
KIRIN M, MCQUILLAN R, FRANKLIN C S, et al. Genomic runs of homozygosity record population history and consanguinity[J]. PLoS One, 2010, 5(11): e13996. DOI:10.1371/journal.pone.0013996 |
| [41] |
LYNCH M, CONERY J, BURGER R. Mutationaccumulation and the extinction of small populations[J]. Am Nat, 1995, 146(4): 489-518. DOI:10.1086/285812 |
| [42] |
CHARLESWORTH D, WILLIS J H. The genetics of inbreeding depression[J]. Nat Rev Genet, 2009, 10(11): 783-796. DOI:10.1038/nrg2664 |
| [43] |
AGRAWAL A F, WHITLOCKM C W. Mutation load: the fitness of individuals in populations where deleterious alleles are abundant[J]. Annu Rev Ecol Evol Syst, 2012, 43(1): 115-135. DOI:10.1146/annurev-ecolsys-110411-160257 |
(编辑 郭云雁)



