2. 农业农村部兽用药物与兽医诊断技术四川科学观测实验站,成都 611130;
3. 四川农业大学国家级动物类实验教学示范中心,成都 611130
2. Sichuan Science-observation Experimental Station of Veterinary Drugs and Veterinary Diagnostic Technology, Ministry of Agriculture and Rural Affairs, Chengdu 611130, China;
3. National Animal Experiments Teaching Demonstration Center, Sichuan Agricultural University, Chengdu 611130, China
丁型冠状病毒(deltacoronavirus)是新发现的冠状病毒科(Coronaviridae),丁型冠状病毒属(Deltacoronavirus)成员,可以感染哺乳动物和禽类[1]。2009年,Woo等[2]发现3种禽丁型冠状病毒:夜莺冠状病毒HKU11、画眉冠状病毒HKU12和文鸟冠状病毒HKU13。2012年,Woo等[3]在猪和鸟群中鉴定出7种新型丁型冠状病毒,对猪冠状病毒HKU15-44和HKU15-155毒株NSP13、S和N基因序列分析显示其与亚洲豹猫冠状病毒相似性高达99.8%,说明PDCoV在野生小型哺乳动物和猪之间可能存在跨种传播。2014年,美国暴发猪丁型冠状病毒(porcine deltacoronavirus,PDCoV)感染[4]。随后,韩国、加拿大、泰国、越南、日本和中国等国家也报道PDCoV引起的仔猪腹泻性疾病[5-6]。当前,已报道的PDCoV毒株全基因组序列相对保守,序列分析表明,PDCoV可能起源于麻雀丁型冠状病毒(sparrow deltacoronavirus,SpCoV)[7]。此外,美国报道的4种新型SpCoV与PDCoV亲缘关系更为密切,表明PDCoV在猪和禽类之间也可能发生跨种传播[8]。
PDCoV是引起猪肠道疾病的主要病原,可感染各年龄阶段的猪,感染仔猪出现呕吐、腹泻、脱水和死亡等临床症状,影响全球养猪业的健康发展[9]。人工接种PDCoV还可感染牛、鸡和火鸡等多种动物[10-12]。体外试验证实,病毒也可感染猪源细胞(LLC-PK、PK15、ST、IPEC和IPI-2I)、人源细胞(Huh7和Hela)、禽源细胞(LMH和DF-1)、牛源细胞(PBK和PBH)、猴源细胞(Vero-CCL81)和犬源细胞(MDCK)等多种细胞,显示PDCoV具有广泛的跨宿主传播风险[7, 13-14]。
氨基肽酶N(aminopeptidase N,APN),又称CD13,是一种膜结合的金属蛋白酶,可与冠状病毒S蛋白结合,介导病毒入侵宿主细胞[15]。甲型冠状病毒中,传染性胃肠炎病毒(transmissible gastroenteritis virus,TGEV)、猫冠状病毒(feline coronavirus,FCoV)、犬冠状病毒(canine coronavirus,CCoV)和人冠状病毒229E(human coronavirus 229E,HCoV-229E)被鉴定出以APN作为功能受体[16-19]。此外,APN作为冠状病毒受体具有种属特异性。有研究表明,HCoV-229E以hAPN作为受体,而不能以猪APN(porcine APN,pAPN)作为受体[20]。猫APN(feline APN,fAPN)可与TGEV、CCoV、HCoV-229E和FCoV结合[17]。但是,APN在PDCoV入侵宿主细胞的过程中是否发挥受体功能存在争议。Li等[21]构建APN基因敲除细胞证明APN是PDCoV感染多种动物细胞的功能受体,病毒可通过S蛋白的S1B结构域与APN的催化区域发生互作。Wang等[22]也证明,pAPN在PDCoV入侵细胞中发挥受体功能。与之相反,部分研究结果却证明,APN不是PDCoV的受体[23-24]。也有研究显示,APN虽不是PDCoV的关键受体,但其能影响PDCoV的早期感染和增殖过程[25]。另外,Stoian等[26]却认为APN是PDCoV感染细胞的一个非必需细胞受体。
为验证hAPN在PDCoV复制中的作用,本研究首先证实PDCoV可感染HEK293细胞,再进一步构建hAPN基因敲除细胞系和hAPN过表达质粒,验证hAPN在PDCoV复制中的作用。通过同源建模和分子对接模拟PDCoV S蛋白与hAPN蛋白的相互作用,为阐述PDCoV的细胞入侵机制和跨种传播提供新的理论依据。
1 材料与方法 1.1 细胞、病毒和主要试剂HEK293、HEK293T和ST细胞由本实验室冻存;PDCoV四川分离株CHN-SC2015(GenBank收录号:MK355396.1)由本实验室分离、鉴定和保存。pCMV-SPORT6-ANPEP质粒由本实验室构建保存;兔抗ACTB多克隆抗体(AC026),HRP-羊抗兔IgG(AS014),HRP-羊抗鼠IgG(AS003):武汉Abclonal公司;鼠抗ANPEP单克隆抗体(sc-166105):Santa Cruz公司;兔抗PDCoV N多克隆抗体由本实验室制备保存。
1.2 PDCoV感染HEK293细胞将PDCoV以MOI=0.1接种60 mm培养皿中的HEK293细胞,37 ℃吸附1 h,PBS洗2遍,加维持液继续培养。于0、12、24、36和48 h取样,通过RT-qPCR和Western blot检测PDCoV感染HEK293细胞后的病毒含量;将PDCoV感染HEK293细胞24 h后的细胞培养物冻融3次后,连续传至4代,通过RT-PCR检测PDCoV在HEK293细胞上的增殖情况,TCID50检测不同代次的病毒滴度。
1.3 hAPN基因敲除细胞系的构建与鉴定参考hAPN基因(NC_000015)序列,利用网络在线工具(http://chopchop.cbu.uib.no/)设计一对sgRNA(表 1)。合成的sgRNA经退火处理,连接到BsmB Ⅰ酶切的线性化载体lenti CRISPR v2,构建同时表达Cas9和sgRNA的慢病毒转移质粒。用Lipofectamine 3000将质粒按重组质粒∶psPAX2∶pMD2.G=5∶3∶2的比例共转染至HEK293T细胞中,48 h后收上清。待T25细胞瓶中HEK293细胞长至50%时,感染慢病毒,36 h后,用含1 μg·mL-1嘌呤霉素(puromycin)的完全培养基进行抗性筛选;有限稀释法挑选hAPN基因敲除细胞系,将其命名为hAPNKO。通过测序、RT-qPCR和Western blot鉴定hAPN在HEK293细胞上的敲除情况。
|
|
表 1 相关引物序列 Table 1 Primer sequences |
按照105·孔-1的细胞数将野生型细胞(hAPNWT) 和敲除细胞(hAPNKO)接种96孔细胞板,24 h后,避光条件下,每孔加入10 μL CCK-8试剂,37 ℃孵育1 h,测定450 nm的吸光度。计算细胞存活率,细胞存活率=[(As-Ab)/(Ac-Ab)]×100%,As:试验孔(hAPNKO细胞孔);Ac:对照孔(hAPNWT细胞孔);Ab:空白孔(培养基)。
1.5 敲除hAPN对PDCoV复制的影响待60 mm细胞培养皿中hAPNWT和hAPNKO长满单层后,PBS洗2遍,将PDCoV以MOI=0.1的剂量感染细胞,37 ℃孵育1 h,弃病毒液,每孔加入4 mL维持液,于24 h收集细胞悬液和蛋白,细胞悬液用RT-qPCR检测M基因转录水平;蛋白样品用Western blot检测N蛋白表达水平。
1.6 过表达hAPN对PDCoV复制的影响针对hAPN基因的CDs区设计引物,经PCR扩增后连接到载体pIRES2-EGFP,构建hAPN过表达真核载体(命名为pIRES2-EGFP-hAPN)。待HEK293细胞长至50%时,用Lipofectamine 3000分别转染4 μg质粒pIRES2-EGFP和pIRES2-EGFP-hAPN至HEK293细胞;转染24 h后,观察绿色荧光蛋白表达情况;收集细胞和细胞蛋白,分别用RT-qPCR和Western blot检测hAPN表达。以MOI为0.1的PDCoV感染转染pIRES2-EGFP-hAPN的HEK293细胞,同时设转染pIRES2-EGFP空载的HEK293细胞为对照,24 h后,收集细胞悬液和蛋白,细胞悬液通过RT-qPCR检测M基因转录水平;蛋白样品通过Western blot检测N蛋白表达水平。
1.7 同源建模与分子对接hAPN蛋白(PDB ID:4FYQ)、PDCoV S蛋白(PDB ID:6B7 N)和HCoV-229E S(PDB ID:6U7H)的整体结构从蛋白质数据库获取(https://www.rcsb.org)[27]。用SWISS-MODEL(https://swissmodel.expasy.org/)手动构建PDCoV S蛋白和HCoV-229E S蛋白的单体结构和受体结构域(receptor binding domain,RBD),用PyMOL对hAPN、PDCoV S和HCoV-229E S蛋白的三维结构进行可视化分析;用MEGA6、ESPrit 3.0(http://espript.ibcp.fr/ESPript/ESPript/index.php)和PyMOL的align功能对PDCoV和HCoV-229E S蛋白的RBD进行序列和结构比对。用分子对接方法模拟PDCoV S1蛋白与hAPN蛋白的相互作用。
1.8 TCID50测定取100 μL病毒液,按照10倍梯度进行倍比稀释,共稀释7个梯度,每个稀释度设置8个重复,同时设置空白对照。待96孔细胞板中的ST细胞长满单层后,PBS洗2遍,加入100 μL稀释好的病毒液,37 ℃孵育1.5 h,弃病毒液,加入150 sμL含5 μg·mL-1胰酶的维持液,37 ℃继续培养。每日观察细胞病变,连续观察4 d,按照Reed & Muench方法计算TCID50。
1.9 统计学分析数据用“平均数±标准差(x±s)”表示,使用GraphPad Prism 5.0软件统计处理,以T检验比较hAPNWT和hAPNKO两组数据间差异,P < 0.05为有统计学意义。
2 结果 2.1 PDCoV在HEK293细胞中的增殖情况将PDCoV以MOI=0.1感染HEK293细胞,收取0、12、24、36和48 h的样品,RT-qPCR检测病毒含量。结果发现,病毒感染细胞12~36 h时,病毒快速增殖,36 h达到顶峰;36~48 h,出现下降趋势(图 1A);此外,在0~24 h,病毒N蛋白表达水平也迅速增加(图 1C)。将PDCoV在HEK293细胞上连续传至4代,RT-PCR检测发现当传至第3代时,只能检测到很弱的条带,而传至第4代时,检测不到条带(图 1B);Western blot检测发现,PDCoV传至2代时,仍可检测到很弱的病毒N蛋白表达水平(图 1D);TCID50结果表明,PDCoV在HEK293细胞上第1代滴度约为4.36 lg TCID50·mL-1,传至2代时,病毒滴度稍微下降,约为3 lg TCID50·mL-1 (图 1E),表明PDCoV可感染HEK293细胞,但是不能在HEK293细胞上稳定传代。
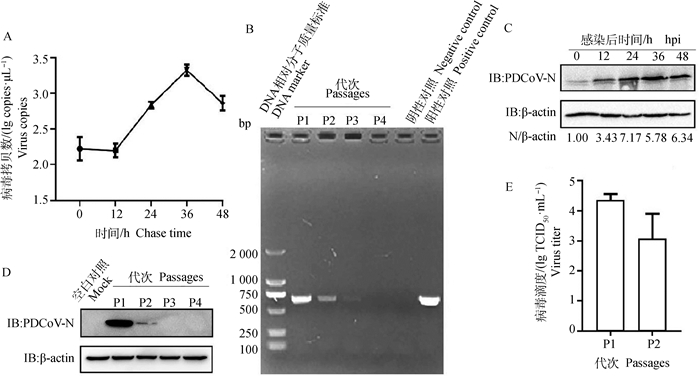
|
A. RT-qPCR检测PDCoV在HEK293细胞上的增殖情况;B. RT-PCR检测PDCoV在HEK293细胞上的传代;C. Western blot检测PDCoV感染细胞不同时间点的N蛋白表达水平;D. Western blot检测PDCoV感染HEK293细胞不同代次的N蛋白表达水平;E. TCID50检测PDCoV感染HEK293细胞不同代次的病毒滴度 A. The proliferation of PDCoV on HEK293 cells was identified by RT-qPCR; B. Detection of PDCoV from different passages on HEK293 cells; C. N protein expression in PDCoV infected cells at different time points was detected by Western blot; D. N protein expression level at different passages from PDCoV infected cells was detected by Western blot; E. Virus titer at different passages from PDCoV infected cells was analyzed by TCID50 assay 图 1 PDCoV在HEK293细胞上的增殖情况 Fig. 1 Propagation of PDCoV on HEK 293 cells |
用CRISPR/Cas9技术构建hAPN基因敲除细胞系,通过测序、RT-qPCR和Western blot检测hAPN在HEK293细胞上的敲除情况。测序结果显示,hAPNKO细胞系在PAM序列前出现明显的重叠峰,且存在23个碱基的连续缺失(图 2A);RT-qPCR和Western blot结果显示,在hAPNKO细胞上几乎检测不到hAPN基因表达(图 2B、C),表明hAPN在HEK293细胞上缺失成功。细胞活性检测发现,hAPNKO和hAPNWT的活性无差异(图 2D),表明敲除hAPN对HEK293细胞活性无影响。
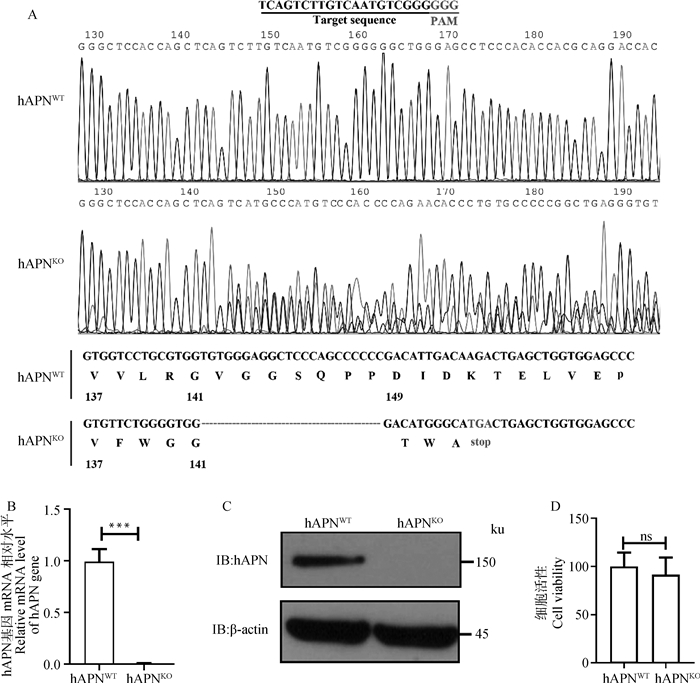
|
A. hAPNWT和hAPNKO细胞中hAPN基因的序列测定;B. RT-qPCR检测hAPNKO细胞的hAPN基因mRNA水平(***.P < 0.001);C. Western blot检测hAPNKO细胞的hAPN蛋白表达水平;D. CCK-8检测hAPNWT和hAPNKO细胞的细胞活性(ns.P>0.05) A. Sequencing of hAPN gene in hAPNWT and hAPNKOcells; B. The relative mRNA level of hAPN gene in hAPNKO cells was confirmed by RT-qPCR(***. P < 0.001); C. The protein expression level of hAPN in hAPNKO cells was confirmed by Western blot; D. Cell viability of hAPNWTand hAPNKO cells was analyzed by CCK-8 (ns. P>0.05) 图 2 hAPN基因敲除细胞系的构建与鉴定 Fig. 2 Construction and identification of hAPN knockout cell lines |
为验证hAPN敲除对PDCoV复制的影响,将PDCoV以MOI=0.1感染hAPNWT和hAPNKO细胞,24 h后,测定M基因转录水平和N蛋白表达水平。结果表明,hAPN基因敲除后,导致M基因mRNA水平下调约75%(图 3 A),N蛋白表达水平下降约66%(图 3 B),证实hAPN敲除能够显著抑制PDCoV复制。
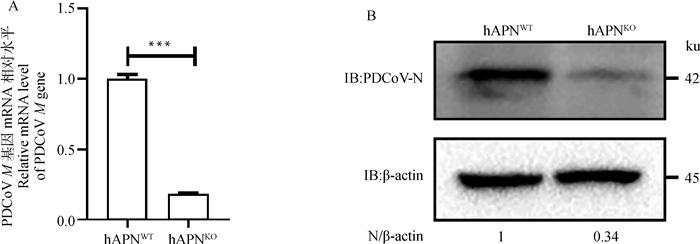
|
A. RT-qPCR检测敲除hAPN后PDCoV M基因mRNA水平(***. P < 0.001);B. Western blot检测敲除hAPN后PDCoV N蛋白表达水平 A. The relative mRNA level of PDCoV M gene was confirmed by RT-qPCR (***. P < 0.001); B. PDCoV N protein expression level was confirmed by Western blot 图 3 敲除hAPN降低PDCoV复制 Fig. 3 Knockout of hAPN reduce PDCoV replication |
为进一步验证hPAN过表达对PDCoV复制的影响,构建hAPN真核表达质粒pIRES2-EGFP-hAPN,转染至HEK293细胞中,24 h后,可以观察到明显的绿色荧光(图 4 A)。RT-qPCR和Western blot检测发现,hAPN在HEK293细胞中成功表达(图 4 B、C)。将pIRES2-EGFP和pIRES2-EGFP-hAPN分别转染HEK293细胞24 h后,接种PDCoV,病毒感染24 h后,RT-qPCR和Western blot检测表明,过表达hAPN导致M基因mRNA水平上调2.7倍,N蛋白表达水平上调1.7倍(图 4 D、E),表明过表达hAPN促进PDCoV复制。
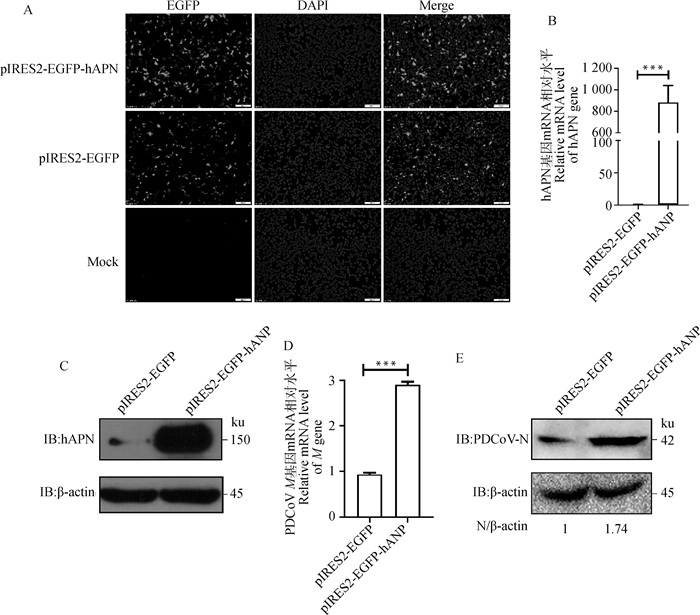
|
A. 荧光显微镜观察绿色荧光蛋白的表达(标尺=100 μm);B. RT-qPCR检测HEK293细胞转染pIRES2-EGFP-hAPN后hAPN基因mRNA水平(***. P < 0.001);C. Western blot检测HEK293细胞转染pIRES2-EGFP-hAPN后hAPN蛋白表达水平;D. RT-qPCR检测过表达hAPN后M基因mRNA水平(***. P < 0.001);E. Western blot检测过表达hAPN后N蛋白表达水平。扫描文章首页OSID码可查看彩图 A. The expression of green fluorescent protein was observed under fluorescence microscope (Bar=100 μm); B. hAPN gene mRNA level was confirmed by RT-qPCR (***. P < 0.001); C. hAPN protein expression level was confirmed by Western blot; D. The relative mRNA level of PDCoV M gene was confirmed by RT-qPCR (***. P < 0.001); E. PDCoV N protein expression level was confirmed by Western blot. The color picture can be found by scanning the OSID code on the front page of the article 图 4 过表达hAPN促进PDCoV复制 Fig. 4 Overexpression of hAPN enhance PDCoV replication |
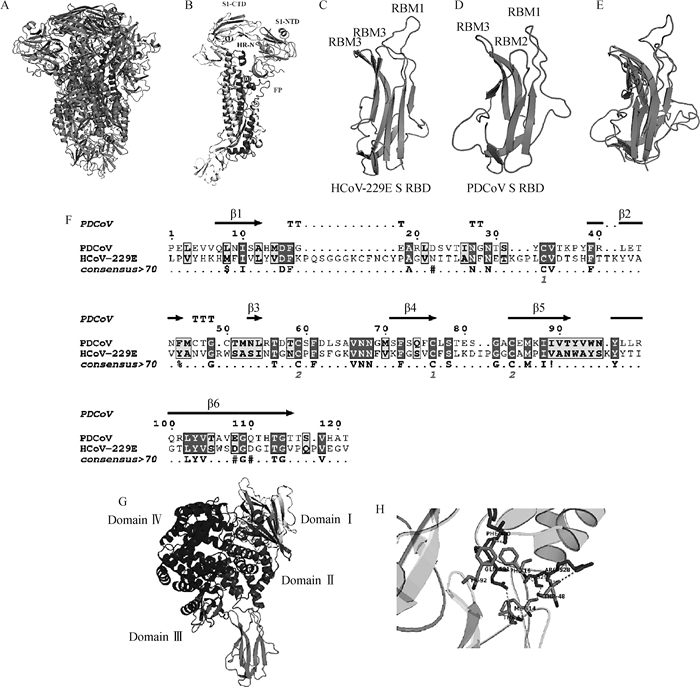
为分析hAPN和PDCoV S蛋白的互作关系,从PDB数据库获取hAPN和PDCoV S的三聚体结构(图 5 A、G);PDCoV S蛋白的单体结构及其主要结构域包括C-末端结构域(C-terminal domain,CTD)、N-末端结构域(N-terminal domain,NTD)、融合肽(fusion peptide,FP)、七肽重复序列(heptad repeat,HR)(图 5 B)。HCoV-229E和PDCoV S1 RBD,包含6个β-折叠和3个短而不连续的环组成的受体结合基序(receptor binding motifs,RBM)(图 5 C、D)。序列和结构比对发现PDCoV和HCoV-229E S1 RBD具有相似的空间结构,表明PDCoV RBD与hAPN的结合可能类似HCoV-229E RBD和hAPN的结合(图 5 E、F)。分子对接结果表明,PDCoV S1 RBD能够结合hAPN结构域Ⅱ,主要是S1蛋白的RBM1氨基酸残基TYR92、THR51、THR48、PHE16和MET14通过氢键与hAPN的氨基酸残基PHE490、GLN531、ARG528和SER529结合(图 5 G、H)。
|
A. PDCoV S蛋白整体结构;B. PDCoV S蛋白单体结构;C. HCoV-229E S1 RBD;D. PDCoV S1 RBD;E. HCoV-229E和PDCoV S1 RBD结构比对;F. HCoV-229E和PDCoV S1 RBD序列比对;G~H. 分子对接模拟PDCoV S1 RBD和hAPN蛋白的互作。扫描文章首页OSID码可查看彩图 A. The overall structure of PDCoV S protein; B. The monomer structure of PDCoV S protein; C. The receptor binding domain of HCoV-229E S1 protein; D. The receptor binding domain of PDCoV S1 protein; E. Structure alignment of HCoV-229E and PDCoV S1 RBD; F. Sequence alignment of PDCoV and HCoV-229E S1 RBD; G-H. Interaction of the PDCoV S protein RBD and hAPN modeled by molecular docking. The color pictures can be found by scanning the OSID code on the front page of the article 图 5 hAPN和PDCoV S蛋白的互作分析 Fig. 5 Interaction of hAPN and PDCoV S protein |
PDCoV是近年来新发现的一种猪冠状病毒,人工接种PDCoV能够感染猪、牛、鸡和火鸡等多种动物。PDCoV感染仔猪后引起明显的腹泻症状[28],而感染小鸡后腹泻症状较轻[12],接种PDCoV的小牛甚至未出现腹泻和其他临床症状[10]。Lednicky等[29]在儿童血清样品中检测到变异的PDCoV,首次报道了PDCoV可感染人。鉴于PDCoV在猪群的广泛流行和跨宿主传播风险,研究宿主细胞蛋白在PDCoV复制中的作用具有重要意义。过表达hAPN、fAPN、pAPN、cAPN显著增加细胞对PDCoV的易感性[21]。APN在不同肠段中的差异表达与PDCoV肠道组织嗜性也密切相关[30]。本研究发现敲除hAPN降低PDCoV复制,而过表达hAPN促进PDCoV复制,表明hAPN是影响PDCoV复制的重要宿主细胞因子,丰富了PDCoV跨种传播和致病机制理论。
S蛋白是冠状病毒入侵宿主细胞的关键蛋白,在病毒感染宿主细胞的过程中,S蛋白构象的变化能促进病毒囊膜与宿主细胞膜的融合。此外,宿主细胞内的酸性环境和蛋白水解酶对S蛋白的活化也是病毒囊膜与宿主细胞膜融合所必需的[31-32]。胰蛋白酶可通过增强细胞与细胞之间的融合而促进PDCoV增殖,但在病毒入侵细胞的过程中不发挥关键作用[33]。此外,PDCoV通过胰蛋白酶介导的细胞膜表面入侵ST和IPI-2I细胞的效率明显高于内吞体途径[34]。HEK293细胞是由剪切过的5型腺病毒DNA转染的人胚肾细胞形成的细胞系[35],HEK293 T细胞是能够稳定表达SV40 T抗原的HEK293衍生细胞系。PEDV可以感染HEK293细胞,且与Vero细胞上的增殖特点相似[36]。此外,黄病毒科成员,日本脑炎病毒(Japanese encephalitis virus,JEV)、寨卡病毒(Zika virus,ZIKV)和黄热病毒(yellow fever virus,YFV)也可感染HEK293T细胞[37]。由于HEK293细胞贴壁能力弱,对胰蛋白酶的耐受程度较弱,本研究在不添加胰蛋白酶的情况下,发现PDCoV可在HEK293细胞至少传至2代,表明PDCoV可不依赖于胰蛋白酶入侵HEK293细胞。
CRISPR/Cas9系统是新发现的一种基因编辑工具,广泛应用于研究宿主因子在病毒复制中的作用[38-39]。近年来,CRISPR/Cas9系统也被应用于筛选调节病毒复制的相关宿主因子[40-41]。关于APN是否为PDCoV入侵宿主细胞的受体存在争议。本研究为验证hAPN在PDCoV毒株CHN-SC2015复制中的作用,通过CRISPR/Cas9技术构建hAPN敲除细胞系,发现hAPN敲除导致PDCoV M基因mRNA水平下调约75%,N蛋白表达水平下降约66%。本研究还通过hAPN过表达验证其对病毒复制的影响,结果发现过表达hAPN可导致PDCoV M基因mRNA水平上调2.7倍,N蛋白表达水平上调1.7倍,证实hAPN可明显促进PDCoV复制。
冠状病毒S蛋白主要由S1和S2结构域组成,其中S1主要负责识别受体,而S2介导宿主细胞膜与病毒囊膜的融合[42]。有研究表明,冠状病毒S1 RBD可以与APN结合,介导病毒入侵宿主细胞。HCoV-229E与hAPN的胞外域Ⅱ结合[19],而TGEV主要与pAPN的胞外域Ⅳ结合[43]。pAPN结构域Ⅶ(581—967 aa)在PEDV复制中发挥重要作用[44]。在PDCoV研究中,Zhu等[25]发现PDCoV S1-CTD蛋白可结合pAPN。本研究通过序列和结构比对发现PDCoV与HCoV-229E S1 RBD具有相似的空间结构,表明PDCoV可能与hAPN结合。因此,本研究通过分子对接方法模拟PDCoV S1 RBD与hAPN的结合,发现PDCoV S1 RBD能够通过RBM1与hAPN结构域Ⅱ结合。
4 结论PDCoV可以感染HEK293细胞,宿主细胞因子hAPN表达可促进PDCoV复制。此外,PDCoV S1 RBD可通过RBM1氨基酸残基TYR92、THR51、THR48、PHE16和MET14与hAPN蛋白的氨基酸残基PHE490、GLN531、ARG528和SER529结合,证实了hAPN是影响PDCoV复制的一个重要宿主细胞因子。
| [1] |
VLASOVA A N, KENNEY S P, JUNG K, et al. Deltacoronavirus evolution and transmission: current scenario and evolutionary perspectives[J]. Front Vet Sci, 2021, 7: 626785. DOI:10.3389/fvets.2020.626785 |
| [2] |
WOO P C Y, LAU S K P, LAM C S F, et al. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus[J]. J Virol, 2009, 83(2): 908-917. DOI:10.1128/JVI.01977-08 |
| [3] |
WOO P C Y, LAU S K P, LAM C S F, et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus[J]. J Virol, 2012, 86(7): 3995-4008. DOI:10.1128/JVI.06540-11 |
| [4] |
WANG L Y, BYRUM B, ZHANG Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014[J]. Emerg Infect Dis, 2014, 20(7): 1227-1230. |
| [5] |
DONG N, FANG L R, ZENG S L, et al. Porcine deltacoronavirus in mainland China[J]. Emerg Infect Dis, 2015, 21(12): 2254-2255. DOI:10.3201/eid2112.150283 |
| [6] |
ZHANG J Q. Porcine deltacoronavirus: overview of infection dynamics, diagnostic methods, prevalence and genetic evolution[J]. Virus Res, 2016, 226: 71-84. DOI:10.1016/j.virusres.2016.05.028 |
| [7] |
WANG Q H, VLASOVA A N, KENNEY S P, et al. Emerging and re-emerging coronaviruses in pigs[J]. Curr Opin Virol, 2019, 34: 39-49. DOI:10.1016/j.coviro.2018.12.001 |
| [8] |
CHEN Q, WANG L Y, YANG C H, et al. The emergence of novel sparrow deltacoronaviruses in the United States more closely related to porcine deltacoronaviruses than sparrow deltacoronavirus HKU17[J]. Emerg Microbes Infect, 2018, 7(1): 105. |
| [9] |
LI B X, ZHENG L L, LI H Y, et al. Porcine deltacoronavirus causes diarrhea in various ages of field-infected pigs in China[J]. Biosci Rep, 2019, 39(9): BSR20190676. DOI:10.1042/BSR20190676 |
| [10] |
JUNG K, HU H, SAIF L J. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus[J]. Arch Virol, 2017, 162(8): 2357-2362. DOI:10.1007/s00705-017-3351-z |
| [11] |
BOLEY P A, ALHAMO M A, LOSSIE G, et al. Porcine deltacoronavirus infection and transmission in poultry, United States[J]. Emerg Infect Dis, 2020, 26(2): 255-265. DOI:10.3201/eid2602.190346 |
| [12] |
LIANG Q Q, ZHANG H L, LI B X, et al. Susceptibility of chickens to porcine deltacoronavirus infection[J]. Viruses, 2019, 11(6): 573. DOI:10.3390/v11060573 |
| [13] |
WANG X L, FANG L R, LIU S D, et al. Susceptibility of porcine IPI-2I intestinal epithelial cells to infection with swine enteric coronaviruses[J]. Vet Microbiol, 2019, 233: 21-27. DOI:10.1016/j.vetmic.2019.04.014 |
| [14] |
JUNG K, VASQUEZ-LEE M, SAIF L J. Replicative capacity of porcine deltacoronavirus and porcine epidemic diarrhea virus in primary bovine mesenchymal cells[J]. Vet Microbiol, 2020, 244: 108660. DOI:10.1016/j.vetmic.2020.108660 |
| [15] |
LIU C, TANG J, MA Y M, et al. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus[J]. J Virol, 2015, 89(11): 6121-6125. DOI:10.1128/JVI.00430-15 |
| [16] |
DELMAS B, GELFI J, L'HARIDON R, et al. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV[J]. Nature, 1992, 357(6377): 417-420. DOI:10.1038/357417a0 |
| [17] |
TRESNAN D B, LEVIS R, HOLMES K V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I[J]. J Virol, 1996, 70(12): 8669-8674. DOI:10.1128/jvi.70.12.8669-8674.1996 |
| [18] |
KOLB A F, HEGYI A, MAILE J, et al. Molecular analysis of the coronavirus-receptor function of aminopeptidase N[M]//ENJUANES L, SIDDELL S G, SPAAN W. Coronaviruses and Arteriviruses. Boston: Springer, 1998: 61-67.
|
| [19] |
YEAGER C L, ASHMUN R A, WILLIAMS R K, et al. Human aminopeptidase N is a receptor for human coronavirus 229E[J]. Nature, 1992, 357(6377): 420-422. DOI:10.1038/357420a0 |
| [20] |
KOLB A F, MAILE J, HEISTER A, et al. Characterization of functional domains in the human coronavirus HCV 229E receptor[J]. J Gen Virol, 1996, 77(10): 2515-2521. DOI:10.1099/0022-1317-77-10-2515 |
| [21] |
LI W T, HULSWIT R J G, KENNEY S P, et al. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility[J]. Proc Natl Acad Sci U S A, 2018, 115(22): E5135-E5143. |
| [22] |
WANG B, LIU Y, JI C M, et al. Porcine deltacoronavirus engages the transmissible gastroenteritis virus functional receptor porcine aminopeptidase N for infectious cellular entry[J]. J Virol, 2018, 92(12): e00318-18. DOI:10.1128/JVI.00318-18 |
| [23] |
卢曼曼, 张家林, 王洪峰, 等. 猪氨基肽酶N不是猪德尔塔冠状病毒入侵宿主细胞的受体[J]. 中国预防兽医学报, 2017, 39(9): 17-22. LU M M, ZHANG J L, WANG H F, et al. Porcine amino peptidase N (pAPN) is not a cellular receptor for porcine deltacoronavirus entry cell[J]. Chinese Journal of Preventive Veterinary Medicine, 2017, 39(9): 17-22. (in Chinese) |
| [24] |
SHANG J, ZHENG Y, YANG Y, et al. Cryo-electron microscopy structure of porcine deltacoronavirus spike protein in the prefusion state[J]. J Virol, 2018, 92(4): e01556-17. DOI:10.1128/JVI.01556-17 |
| [25] |
ZHU X Y, LIU S D, WANG X L, et al. Contribution of porcine aminopeptidase N to porcine deltacoronavirus infection[J]. Emerg Microbes Infect, 2018, 7(1): 65. |
| [26] |
STOIAN A, ROWLAND R R R, PETROVAN V, et al. The use of cells from ANPEP knockout pigs to evaluate the role of aminopeptidase N (APN) as a receptor for porcine deltacoronavirus (PDCoV)[J]. Virology, 2020, 541: 136-140. DOI:10.1016/j.virol.2019.12.007 |
| [27] |
BERMAN H M, WESTBROOK J, FENG Z K, et al. The protein data bank[J]. Nucl Acids Res, 2000, 28(1): 235-242. DOI:10.1093/nar/28.1.235 |
| [28] |
DONG N, FANG L R, YANG H, et al. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014[J]. Vet Microbiol, 2016, 196: 98-106. DOI:10.1016/j.vetmic.2016.10.022 |
| [29] |
LEDNICKY J A, TAGLIAMONTE M S, WHITE S K, et al. Independent infections of porcine deltacoronavirus among Haitian children[J]. Nature, 2021, 600(7887): 133-137. DOI:10.1038/s41586-021-04111-z |
| [30] |
YIN L D, CHEN J F, LI L, et al. Aminopeptidase N expression, not interferon responses, determines the intestinal segmental tropism of porcine deltacoronavirus[J]. J Virol, 2020, 94(14): e00480-20. DOI:10.1128/JVI.00480-20 |
| [31] |
WICHT O, LI W T, WILLEMS L, et al. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture[J]. J Virol, 2014, 88(14): 7952-7961. DOI:10.1128/JVI.00297-14 |
| [32] |
SHIRATO K, KANOU K, KAWASE M, et al. Clinical isolates of human coronavirus 229E bypass the endosome for cell entry[J]. J Virol, 2017, 91(1): e01387-16. DOI:10.1128/JVI.01387-16 |
| [33] |
YANG Y L, MENG F D, QIN P, et al. Trypsin promotes porcine deltacoronavirus mediating cell-to-cell fusion in a cell type-dependent manner[J]. Emerg Microbes Infect, 2020, 9(1): 457-468. DOI:10.1080/22221751.2020.1730245 |
| [34] |
ZHANG J L, CHEN J F, SHI D, et al. Porcine deltacoronavirus enters cells via two pathways: a protease-mediated one at the cell surface and another facilitated by cathepsins in the endosome[J]. J Biol Chem, 2019, 294(25): 9830-9843. DOI:10.1074/jbc.RA119.007779 |
| [35] |
STEPANENKO A A, DMITRENKO V V. HEK293 in cell biology and cancer research: phenotype, karyotype, tumorigenicity, and stress-induced genome-phenotype evolution[J]. Gene, 2015, 569(2): 182-190. DOI:10.1016/j.gene.2015.05.065 |
| [36] |
ZHANG J, GUO L J, XU Y F, et al. Characterization of porcine epidemic diarrhea virus infectivity in human embryonic kidney cells[J]. Arch Virol, 2017, 162(8): 2415-2419. DOI:10.1007/s00705-017-3369-2 |
| [37] |
ZHANG R, MINER J J, GORMAN M J, et al. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses[J]. Nature, 2016, 535(7610): 164-168. DOI:10.1038/nature18625 |
| [38] |
MAR K B, RINKENBERGER N R, BOYS I N, et al. LY6E mediates an evolutionarily conserved enhancement of virus infection by targeting a late entry step[J]. Nat Commun, 2018, 9(1): 3603. DOI:10.1038/s41467-018-06000-y |
| [39] |
MA L, LI F, ZHANG J W, et al. Host factor SPCS1 regulates the replication of japanese encephalitis virus through interactions with transmembrane domains of NS2B[J]. J Virol, 2018, 92(12): e00197-18. DOI:10.1128/JVI.00197-18 |
| [40] |
ZHAO C Z, LIU H L, XIAO T H, et al. CRISPR screening of porcine sgRNA library identifies host factors associated with Japanese encephalitis virus replication[J]. Nat Commun, 2020, 11(1): 5178. DOI:10.1038/s41467-020-18936-1 |
| [41] |
FLINT M, CHATTERJEE P, LIN D L, et al. A genome-wide CRISPR screen identifies N-acetylglucosamine-1-phosphate transferase as a potential antiviral target for Ebola virus[J]. Nat Commun, 2019, 10(1): 285. DOI:10.1038/s41467-018-08135-4 |
| [42] |
QIAN Z H, OU X Y, GÓES L G B, et al. Identification of the receptor-binding domain of the spike glycoprotein of human betacoronavirus HKU1[J]. J Virol, 2015, 89(17): 8816-8827. DOI:10.1128/JVI.03737-14 |
| [43] |
DELMAS B, GELFI J, KUT E, et al. Determinants essential for the transmissible gastroenteritis virus-receptor interaction reside within a domain of aminopeptidase-N that is distinct from the enzymatic site[J]. J Virol, 1994, 68(8): 5216-5224. DOI:10.1128/jvi.68.8.5216-5224.1994 |
| [44] |
KAMAU A N, PARK J E, PARK E S, et al. Porcine amino peptidase N domain Ⅶ has critical role in binding and entry of porcine epidemic diarrhea virus[J]. Virus Res, 2017, 227: 150-157. DOI:10.1016/j.virusres.2016.10.004 |
(编辑 白永平)



