2. 新疆生产建设兵团塔里木畜牧科技重点实验室,阿拉尔 843300
2. Key Laboratory of Tarim Animal Husbandry Science and Technology, Xinjiang Production and Construction Corps, Alaer 843300, China
新疆南疆和田地区位于塔克拉玛干沙漠南缘,昆仑山北麓。年降水量在21~146 mm,年均蒸发量达2 600 mm,每年沙尘天气220 d以上,其中浓浮尘(沙尘暴)天气在60 d以上,生态环境脆弱。该地区汉朝已有养羊业记载,地方品种绵羊与和田羊、策勒黑羊两个品种的相同点:鼻梁微隆,耳大、下垂,体格较小,舍饲状态常年发情。不同点:和田羊被毛白色,以产地毯毛为主;策勒黑羊被毛黑色,以产羔皮和多胎著称。经过长期选择能抵抗多变的沙漠环境,是我国独特的优势种质资源绵羊群体。
家畜在历史驯化和品种培育及改良过程中受到强烈的自然选择和人工选择,在家畜基因组上留下相应的遗传印记,具体表现为基因组上连锁不平衡程度增加或降低和受选择位点及其附近遗传多样性降低。2004年美国、英国和新西兰的科学家公布了第三代高密度的绵羊遗传连锁图[1]。绵羊基因组学的研究是开展绵羊分子遗传标记研究与应用的基础[2-3],2014年,华大基因和中国科学院的研究者公布了绵羊基因组序列,以雌性陶赛特为绵羊材料,通过二代测序技术与基因组图谱技术相结合,测序深度为75X,其中Contig N50为39 959 bp,Scaffold N50为2 231 873 bp,共包括20 908个基因,141个大结构改变,近一万个拷贝数变异,大约有1 000万个SNPs,基因组全长2.61 Gb[4]。绵羊基因组测序的完善为功能基因定位、相关分子标记的研究提供了遗传信息。Li等[5]选择99只Finnsheep不同颜色的绵羊群体,通过全基因组关联分析和选择性清扫相结合的方法,获得了毛色相关的基因(TYRP、ASIP、MITF)。Kijas等[6]基于Ovine SNP50 BeadChip通过Fst方法对全世界74个绵羊品种进行了品种间的选择性清扫检测,共获得181个基因,其中KIT、MITF和色素沉积相关;ASIP和黑、白色或黑白相间的毛色相关;NPR2、HMGA2、BMP2和动物的身体大小、肌肉肥大等生产性状相关;FGF5和毛质性状相关等。Zhao等[7]用3个典型的中国本土绵羊品种,基于Ovine Infinium HD SNP BeadChip进行选择性清扫,其中25个基因组区域被选择,筛选出73个候选基因,这些基因与尾部脂肪的形成、繁殖、生长性状等有关。沙漠环境抗逆性是在沙漠极端环境下生物生长发育和生产性能表现正常的能力,外来引入品种对沙漠环境适应性差[8],生产性能下降[9],Yang等[10]使用重测序方法从不同的生态环境中获得77个绵羊基因组,并确定绵羊与沙漠(ANXA6、GPX3、GPX7和PTGS2)和高原(IFNGR2、MAPK4、NOX4、SLC2A4和PDK1)等极端环境相适应有关的相关基因。
和田地区存在和田羊和策勒黑羊两个地方绵羊品种,不断的自然选择和人工选择在这两种绵羊基因组上留下了大量的遗传印记。和田羊与策勒黑羊均具有适应沙漠环境的能力,但是关于两个品种的基因组差异研究较少。本研究旨在利用基因组选择性清扫检测和田羊和策勒黑羊的基因组差异,寻找适应南疆沙漠环境的优异分子标记,为南疆土著绵羊保护和育种提供理论基础。
1 材料与方法 1.1 样品采集本试验于和田地区策勒县绿源种畜场随机选取策勒黑羊41只,于和田羊保种场随机选取和田羊84只,均为成年母羊,采样时间为2018—2021年。藏绵羊37只,数据来自International Stroke Genetics Consortium (ISGC)(http://www.Sheephapmap.org)。
1.2 DNA提取和基因组芯片制备利用耳缺钳在每个个体的耳缘处剪下直径3 mm的组织块,放入己灭菌含有75%酒精的离心管。采用苯酚-氯仿法提取DNA,TAE电泳和核酸仪检测分析后,进行Ilumina Ovine SNP50 BeadChip制备。BeadScan软件对图像进行转换,利用Genome Studio软件对数据结果进行处理。
1.3 质控分析采用Plink1.07[11]软件进行数据质控,质控标准为:去掉样本中检出率小于95%的个体样品,SNPs检出率小于90%、最小等位基因频率小于5%、哈代-温伯格平衡检验P值小于1×10-6。
1.4 主成分分析使用GCTA软件[12]对过滤后的SNPs数据进行PCA分析,该主成分分析基于个体间的遗传相关性识别,代表种群结构的主要成分[13]。
1.5 基因组选择信号的计算本试验以和田羊和策勒黑羊为研究对象,采用HapFLK方法,选择藏绵羊作为外群。HapFLK软件[14]基于群体间单体型频率来检测选择差异。使用Arlequin软件[15]来构建雷诺(Reynolds)亲属矩阵,在fastPHASE模型中,亲缘关系矩阵假设有10个群集(cluster),使用20次迭代的期望最大化算法(EM)对所有染色体进行FLK分析。得出结果后,利用以下公式校正HapFLK统计量:
| $ {{\rm{hapFL}}{{\rm{K}}_{{\rm{adj}}}} = \frac{{{\rm{hapFLK}} - {\rm{mean}}\left( {{\rm{hapFLK}}} \right)}}{{{\rm{sd}}\left( {{\rm{hapFLK}}} \right)}}} $ |
其中,mean(hapFLK)为hapFLK统计量的平均数,sd(hapFLK)为hapFLK统计量的标准偏差。校正以后的HapFLK统计量近似服从正态分布,利用Excel 2016的函数1-NORM.DIST(HapFLKadj, 0, 1, TRUE)得到每个位点的P值, 若-lgP≥3认为该位点受到了选择[16]。
1.6 GO和KEGG富集分析所有获得SNPs基于Oar_v4.0 (https://www.sheephapmap.org/)进行注释。基因的功能分析参考NCBI数据库(http:/www.ncbi.nlm.nih.gov/gene)、OMIM数据库(http://www.ncbi.nlm.nih.gov/omim)以及相应的参考文献。使用DAVID工具[17](http://david.Abcc.ncifcrf.gov/)进行GO和KEGG (Kyoto Encyclopedia of Genes and Genomes)分析。每个富集条目的重要程度由P值衡量,P值遵循超几何分布,公式为:
| $ {P = 1 - \sum\limits_{i = 0}^{m - i} {\frac{{\left( {\frac{M}{i}} \right)\left( {\frac{{N - M}}{{n - i}}} \right)}}{{\left( {\frac{N}{n}} \right)}}} } $ |
其中,N为总基因数;n为N中差异表达基因的总数;M为N中属于某个富集条目的基因个数;i为n中属于某个富集条目的基因个数。为了避免结果假阳性,对富集分析结果P采用多重检验(False Discovery Rate, FDR)进行校正。FDR的公式为:
| $ {{\rm{FDR}} = P \times \frac{n}{{(rankP)}}} $ |
其中,P是原始P值,n是测试次数,rankP是特定原始P值的等级。FDR≤0.05时,符合此条件的GO术语和途径被定义为候选基因显著富集。
2 结果 2.1 主成分分析如图 1所示,藏羊与策勒黑羊、和田羊群体可以由第一主成分分为两组,前两个主成分(PC1和PC2)可以将所有样本分为4个子组,PC2与PC3两个主成分也大致将所有样本分为4个组。其中藏羊群体与和田羊、策勒黑羊群体明显区分为两个群体,和田羊群体与策勒黑羊群体基因组差异很小。

|
A、B、C分别为PC1-PC2、PC1-PC3和PC2-PC3所对应的区分结果;PCA结果将所有个体大致分为3组,其中和田羊与策勒黑羊遗传关系较近 A, B and C are the differentiation results corresponding to PC1-PC2, PC1-PC3 and PC2-PC3, respectively; the PCA results roughly divide all individuals into 3 groups, the Hetian and Qira Black sheep have more close genetic relationship 图 1 3个绵羊群体的主成分分析图 Fig. 1 Principal component analysis plots for the 3 sheep populations |
采用HapFLK结果的前1%作为有效结果,共计470个SNPs,结果如图 2所示,OAR1、OAR3、OAR6号染色体上的选择信号较强烈。通过基因注释(距离该位点最近的基因)找到这些位点相对应的候选基因,并去除重复与无效的基因,共得到了167个基因,其中绵羊毛生长相关基因有KRT82、KRT80、KRT1、KRT79、KRT8、KRT7、KRT77、KRT5、KRT84、KRT18;毛色相关基因为KIT,生长发育相关基因有AMHR2、SP1、OARDACVR1B、ADIPOQ。候选位点及注释结果见表 1。

|
黑色虚线表示显著水平,黑色实线表示极显著水平 The black dashed line indicates significant level and the black solid line indicates highly significant level 图 2 各染色体SNP位点对应的HapFLK统计量 Fig. 2 The HapFLK statistic corresponding to each chromosomal SNP locus |
|
|
表 1 HapFLK候选位点及其基因 Table 1 HapFLK candidate sites and their genes |
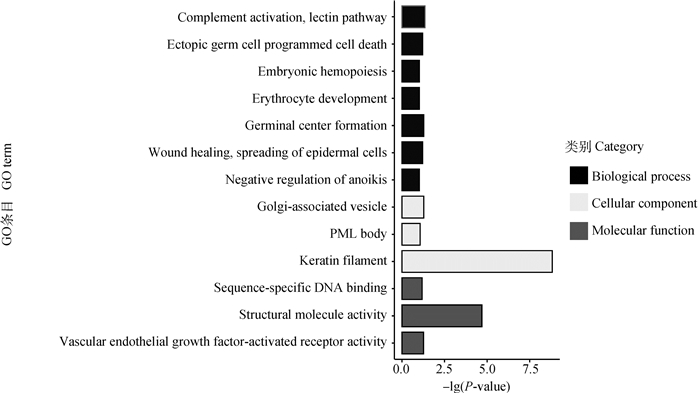
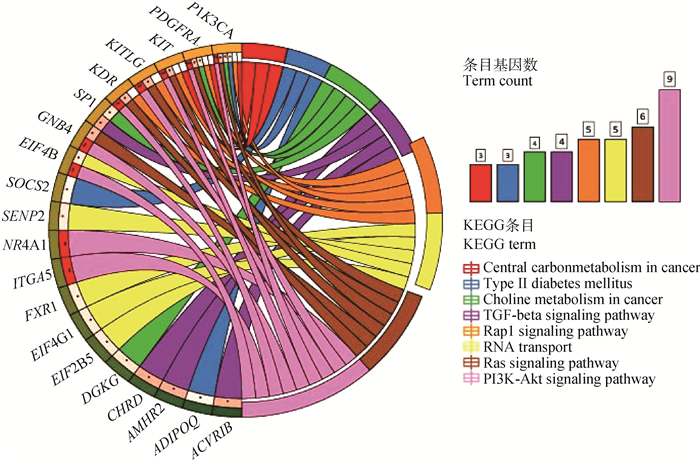
将选择信号突出的1、3、6号染色体的候选基因进行富集分析,内容包括细胞组分(cellular component)、分子功能(molecular function)、生物学过程(biological process)和KEGG信号通路分析。167个候选基因参与富集,对P值进行FDR校正后,共得到7个显著GO组分和6个显著KEGG通路。候选基因GO分析显著结果如表 2所示,每个条目名称及其数量见图 3;候选基因KEGG通路及其关系见图 4。
|
|
表 2 候选基因显著富集的GO条目 Table 2 The significant GO terms enriched by candidate genes |
|
图 3 候选基因富集分析得到的GO条目及其基因数 Fig. 3 GO term and their gene counts obtained from candidate genes enrichment analysis |
|
图 4 候选基因KEGG通路及其相互关系 Fig. 4 Candidate genes KEGG pathway and their relationship |
通过基因组选择性清扫检测物种的起源进化和基因交流[18],探究家畜在环境变化过程中的遗传机理。不同绵羊品种之间分子遗传标记不同,利用基因组选择性清扫的方法获得具有一定意义的分子标记。本研究采用HapFLK方法,以藏绵羊作为外群,在剔除祖先成分之后,选择性清扫结果集中体现了两个品种间的基因组差异。两个绵羊品种的主要差异体现在毛质、毛色、尾型等性状,这些结果在表型上也有体现。在表型上,和田羊与策勒黑羊在全年发情及多胎性上不一致,但在基因组清扫中并未体现其差异,认为和田羊与策勒黑羊类似,均存在多胎潜力,可能因为饲养方式不同,而没有表现出多胎性状,这个结论与王琼等[19]的研究结果相似。
3.2 角蛋白基因家族基因组选择信号分析获得了KRT82、KRT80、KRT1、KRT79、KRT8、KRT7、KRT77、KRT5、KRT84、KRT18等角蛋白基因家族基因,富集分析结果显示,KRT基因数量较多,且富集程度高。KRT基因主要为角蛋白基因和角蛋白相关基因[20],角蛋白是羊毛纤维的主要成分,是形成皮肤毛囊细胞的主要结构蛋白质,是维持毛囊结构并在毛囊中表达最丰富的蛋白质,在很大程度上决定了羊毛的结构特性,其编码基因是毛囊基因表达和毛发生物学研究中重要的候选基因。该基因家族广泛存在于绵羊的毛发和皮肤中,调节与控制绵羊毛囊及毛发的发育。同时,KRT基因参与动物皮肤组织分化和绵羊毛长、弯曲度、卷曲方式[21]和纤维直径[22-24]调控,是羊毛质量[25-26]的主要调控基因。
经过长时间的选育,两个绵羊品种毛质性状存在很大差异,在生产性状上策勒黑羊被毛为异质毛,而和田羊为地毯型毛。相关研究显示,在绵羊机体中,KRT基因的突变会导致羊毛分叉,变细,变秃。在其他生物中,KRT基因也广泛参与了其相关表型的表达与调控[20, 27-28],在人类的大部分皮肤病中都能找到KRT基因的影子,如人类KRT1突变可能会导致表皮溶解性鱼鳞病(EI)的发生[29],另外,人类的头发相关表型也与KRT基因有关[30-32]。KRT18参与到小鼠肌肉的功能表达和小鼠表皮细胞的分化[33]。KRT7从头表达导致上皮细胞可塑性和干细胞特性的改变是增加终末期肾肿瘤发生风险的关键因素[34]。
3.3 其他的基因及其功能血小板衍生生长因子受体ɑ(platelet derived growth factor receptor alpha, PDGFRA)位于OAR6染色体,物理位置为69 735 803~69 771 932 bp,该基因在机体器官发育、伤口愈合中起着重要的作用,其突变位点与特发性嗜酸性粒细胞增多症和家族性胃肠道间质瘤以及其他多种癌症有关,并参与皮肤及毛发色素沉着的表达[35-36],是绵羊抗逆性和毛发颜色研究的候选基因。
KIT原癌基因,受体酪氨酸激酶(KIT proto-oncogene, receptor tyrosine kinase, KIT)位于OAR6染色体,物理位置为70 189 729~70 234 612 bp。该蛋白磷酸化多种细胞内蛋白,这些蛋白与多种不同类型的细胞增殖、分化、迁移和凋亡相关,在造血、干细胞维持中起重要作用。KIT是调节黑色素生成、控制黑色素细胞增殖和凋亡的关键基因,是影响动物毛皮色素沉着机制的重要基因[37]。和田羊被毛为白色,策勒黑羊被毛为黑色,该基因可能与两个绵羊品种毛色相关。此外,KIT基因是生物环境适应性相关基因,参与绵羊的环境适应性及驯化[38-39]。
激酶插入域受体(kinase insert domain receptor, KDR)位于OAR6染色体,物理位置为70 607 356~ 70 562 533 bp。该基因为血管发育中至关重要的蛋白质的编码基因[40],与类风湿性关节炎(RA) 的易感性[41]和原发性乳腺血管肉瘤的发生[42]紧密相关。
脂联素,C1Q和胶原结构域(adiponectin, C1Q and collagen domain containing, ADIPOQ) 位于OAR1染色体,物理位置为198 631 126~19 861 993 bp,脂联素蛋白基因完全在脂肪组织中表达[43-44]。编码蛋白在血浆中循环,并参与代谢和荷尔蒙生成的过程。该基因极有可能与绵羊尾部脂肪的大小相关,绵羊尾部脂肪是绵羊能量的储存库,具有供能与保暖的作用,是绵羊适应恶劣环境的一个重要特征。该基因可以作为培育适应南疆环境绵羊的候选基因。
4 结论本研究通过基因组选择性清扫的方法,解析和田羊和策勒黑羊的遗传规律和基因组差异,发现两个绵羊品种主要在毛质性状上存在差异,繁殖相关性状差异较小,这表明和田羊经过纯繁能够显著提高繁殖性能。该结果可为南疆绵羊品种的遗传改良提供理论基础和分子选育方向。
| [1] |
CRAWFORD A M, DODDS K G, EDE A J, et al. An autosomal genetic linkage map of the sheep genome[J]. Genetics, 1995, 140(2): 703-724. DOI:10.1093/genetics/140.2.703 |
| [2] |
ROMANOV M N, ZINOVIEVA N A, GRIFFIN D K. British sheep breeds as a part of world sheep gene pool landscape: looking into genomic applications[J]. Animals (Basel), 2021, 11(4): 994. |
| [3] |
MASTRANGELO S, PORTOLANO B, DI GERLANDO R, et al. Genome-wide analysis in endangered populations: a case study in Barbaresca sheep[J]. Animal, 2017, 11(7): 1107-1116. DOI:10.1017/S1751731116002780 |
| [4] |
JIANG Y, XIE M, CHEN W B, et al. The sheep genome illuminates biology of the rumen and lipid metabolism[J]. Science, 2014, 344(6188): 1168-1173. DOI:10.1126/science.1252806 |
| [5] |
LI M, TⅡRIKKA T, KANTANEN J. A genome-wide scan study identifies a single nucleotide substitution in ASIP associated with white versus non-white coat-colour variation in sheep (Ovis aries)[J]. Heredity (Edinb), 2014, 112(2): 122-131. DOI:10.1038/hdy.2013.83 |
| [6] |
KIJAS J W, LENSTRA J A, HAYES B, et al. Genome-wide analysis of the world's sheep breeds reveals high levels of historic mixture and strong recent selection[J]. PLoS Biol, 2012, 10(2): e1001258. DOI:10.1371/journal.pbio.1001258 |
| [7] |
ZHAO F P, DENG T Y, SHI L Y, et al. Genomic scan for selection signature reveals fat deposition in Chinese indigenous sheep with extreme tail types[J]. Animals (Basel), 2020, 10(5): 773. |
| [8] |
CHEDID M, JABER L S, GIGER-REVERDIN S, et al. Review: Water stress in sheep raised under arid conditions[J]. Can J Anim Sci, 2014, 94(2): 243-257. DOI:10.4141/cjas2013-188 |
| [9] |
BERGER J. Female breeding age and lamb survival in desert bighorn sheep (Ovis canadensis)[J]. Mammalia, 1982, 46(2): 183-190. |
| [10] |
YANG J, LI W R, LV F H, et al. Whole-genome sequencing of native sheep provides insights into rapid adaptations to extreme environments[J]. Mol Biol Evol, 2016, 33(10): 2576-2592. DOI:10.1093/molbev/msw129 |
| [11] |
CHANG C C, CHOW C C, TELLIER L C, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets[J]. GigaScience, 2015, 4(1): 7. DOI:10.1186/s13742-015-0047-8 |
| [12] |
YANG J, LEE S H, GODDARD M E, et al. Genome-Wide Complex Trait Analysis (GCTA): methods, data analyses, and interpretations[M]//GONDRO C, VAN DER WERF J, HAYES B. Genome-Wide Association Studies and Genomic Prediction. Totowa: Humana Press, 2013.
|
| [13] |
YUAN Z H, LI W H, LI F D, et al. Selection signature analysis reveals genes underlying sheep milking performance[J]. Arch Anim Breed, 2019, 62(2): 501-508. DOI:10.5194/aab-62-501-2019 |
| [14] |
KIJAS J W. Haplotype-based analysis of selective sweeps in sheep[J]. Genome, 2014, 57(8): 433-437. DOI:10.1139/gen-2014-0049 |
| [15] |
EXCOFFIER L, LAVAL G, SCHNEIDER S. Arlequin (version 3.0): An integrated software package for population genetics data analysis[J]. Evol Bioinform Online, 2005(1): 47-50. |
| [16] |
KOMINAKIS A, TARSANI E, HAGER-THEODORIDES A L, et al. Genetic differentiation of mainland-island sheep of Greece: Implications for identifying candidate genes for long-term local adaptation[J]. PLoS One, 2021, 16(9): e0257461. DOI:10.1371/journal.pone.0257461 |
| [17] |
HUANG D W, SHERMAN B T, TAN Q, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists[J]. Nucleic Acids Res, 2007, 35(S1): W169-W175. |
| [18] |
STEPHAN W. Selective sweeps[J]. Genetics, 2019, 211(1): 5-13. DOI:10.1534/genetics.118.301319 |
| [19] |
王琼, 马海玉, 刘玲玲, 等. GDF9、BMPR1B与BMP15基因在和田羊群体的遗传特征分析[J]. 草食家畜, 2020(4): 1-8. WANG Q, MA H Y, LIU L L, et al. Analysis on the Genetic Characteristics of GDF9, BMPR1B and BMP15 gene in Hotan sheep population[J]. Grass-feeding Livestock, 2020(4): 1-8. (in Chinese) |
| [20] |
ZHANG X W, YIN M M, ZHANG L J. Keratin 6, 16 and 17-critical barrier alarmin molecules in skin wounds and psoriasis[J]. Cells, 2019, 8(8): 807. DOI:10.3390/cells8080807 |
| [21] |
KANG X L, LIU Y F, ZHANG J B, et al. Characteristics and expression profile of KRT71 screened by suppression subtractive hybridization cDNA library in curly fleece Chinese tan sheep[J]. DNA Cell Biol, 2017, 36(7): 552-564. DOI:10.1089/dna.2017.3718 |
| [22] |
ZHAO M L, ZHOU H T, LUO Y Z, et al. Variation in a newly identified caprine KRTAP gene is associated with raw cashmere fiber weight in Longdong cashmere goats[J]. Genes (Basel), 2021, 12(5): 625. DOI:10.3390/genes12050625 |
| [23] |
ZHAO M L, ZHOU H T, LUO Y Z, et al. Variation in the caprine keratin-associated protein 27-1 gene is associated with cashmere fiber diameter[J]. Genes (Basel), 2020, 11(8): 934. DOI:10.3390/genes11080934 |
| [24] |
WANG J Q, ZHOU H T, HICKFORD J G H, et al. Identification of caprine KRTAP28-1 and its effect on cashmere fiber diameter[J]. Genes (Basel), 2020, 11(2): 121. DOI:10.3390/genes11020121 |
| [25] |
SULAYMAN A, TURSUN M, SULAIMAN Y, et al. Association analysis of polymorphisms in six keratin genes with wool traits in sheep[J]. Asian-Australas J Anim Sci, 2018, 31(6): 775-783. DOI:10.5713/ajas.17.0349 |
| [26] |
MA G W, CHU Y K, ZHANG W J, et al. Polymorphisms of FST gene and their association with wool quality traits in Chinese Merino sheep[J]. PLoS One, 2017, 12(4): e0174868. DOI:10.1371/journal.pone.0174868 |
| [27] |
LIN J B, FENG Z, QIU M L, et al. KRT 15 as a prognostic biomarker is highly expressed in esophageal carcinoma[J]. Future Oncol, 2020, 16(25): 1903-1909. DOI:10.2217/fon-2019-0603 |
| [28] |
LIU J, KUMAR S, DOLZHENKO E, et al. Molecular characterization of the transition from acute to chronic kidney injury following ischemia/reperfusion[J]. JCI Insight, 2017, 2(18): e94716. DOI:10.1172/jci.insight.94716 |
| [29] |
JI Y Z, BAI Y, WANG S, et al. A KRT1 gene mutation related to epidermolytic ichthyosis in a Chinese family[J]. Clin Exp Dermatol, 2015, 40(8): 879-882. DOI:10.1111/ced.12649 |
| [30] |
LIU F, CHEN Y, ZHU G, et al. Meta-analysis of genome-wide association studies identifies 8 novel loci involved in shape variation of human head hair[J]. Hum Mol Genet, 2018, 27(3): 559-575. DOI:10.1093/hmg/ddx416 |
| [31] |
OKA A, TAKAGI A, KOMIYAMA E, et al. Alopecia areata susceptibility variant in MHC region impacts expressions of genes contributing to hair keratinization and is involved in hair loss[J]. EBioMedicine, 2020, 57: 102810. DOI:10.1016/j.ebiom.2020.102810 |
| [32] |
ZHANG Z G, WANG X L, ZHANG R Q. Pathogenesis of alopecia areata based on bioinformatics analysis[J]. Indian J Dermatol, 2019, 64(1): 1. |
| [33] |
GUNNARSSON A P, CHRISTENSEN R, LI J, et al. Dataset on gene expression profiling of multiple murine hair follicle populations[J]. Data Brief, 2016, 9: 328-334. DOI:10.1016/j.dib.2016.08.063 |
| [34] |
SARLOS D P, PETERFI L, SZANTO A, et al. Shift of keratin expression profile in end-stage kidney increases the risk of tumor development[J]. Anticancer Res, 2018, 38(9): 5217-5222. DOI:10.21873/anticanres.12845 |
| [35] |
ILLA S K, MUKHERJEE S, NATH S, et al. Genome-wide scanning for signatures of selection revealed the putative genomic regions and candidate genes controlling milk composition and coat color traits in sahiwal cattle[J]. Front Genet, 2021, 12: 699422. DOI:10.3389/fgene.2021.699422 |
| [36] |
TRAN K B, GIMENEZ G, TSAI P, et al. Genomic and signalling pathway characterization of the NZM panel of melanoma cell lines: A valuable model for studying the impact of genetic diversity in melanoma[J]. Pigment Cell Melanoma Res, 2021, 34(1): 136-143. DOI:10.1111/pcmr.12908 |
| [37] |
HU S S, CHEN Y, ZHAO B H, et al. KIT is involved in melanocyte proliferation, apoptosis and melanogenesis in the Rex Rabbit[J]. PeerJ, 2020, 8: e9402. DOI:10.7717/peerj.9402 |
| [38] |
YUAN Z H, ZHANG J X, LI W H, et al. Association of polymorphisms in candidate genes with the litter size in two sheep breeds[J]. Animals (Basel), 2019, 9(11): 958. |
| [39] |
YURCHENKO A A, DENISKOVA T E, YUDIN N S, et al. High-density genotyping reveals signatures of selection related to acclimation and economically important traits in 15 local sheep breeds from Russia[J]. BMC Genomics, 2019, 20(S3): 294. DOI:10.1186/s12864-019-5537-0 |
| [40] |
HEINOLAINEN K, KARAMAN S, D'AMICO G, et al. VEGFR3 modulates vascular permeability by controlling VEGF/VEGFR2 signaling[J]. Circ Res, 2017, 120(9): 1414-1425. DOI:10.1161/CIRCRESAHA.116.310477 |
| [41] |
PARADOWSKA-GORYCKA A, STYPINSKA B, PAWLIK A, et al. KDR (VEGFR2) genetic variants and serum levels in patients with rheumatoid arthritis[J]. Biomolecules, 2019, 9(8): 355. DOI:10.3390/biom9080355 |
| [42] |
BECA F, KRINGS G, CHEN Y Y, et al. Primary mammary angiosarcomas harbor frequent mutations in KDR and PIK3CA and show evidence of distinct pathogenesis[J]. Mod Pathol, 2020, 33(8): 1518-1526. DOI:10.1038/s41379-020-0511-6 |
| [43] |
LI B J, QIAO L Y, AN L X, et al. Transcriptome analysis of adipose tissues from two fat-tailed sheep breeds reveals key genes involved in fat deposition[J]. BMC Genomics, 2018, 19(1): 338. DOI:10.1186/s12864-018-4747-1 |
| [44] |
AN Q M, ZHOU H T, HU J, et al. Haplotypes of the ovine adiponectin gene and their association with growth and carcass traits in New Zealand Romney Lambs[J]. Genes (Basel), 2017, 8(6): 160. DOI:10.3390/genes8060160 |
(编辑 郭云雁)



