牛肉具有高蛋白、低脂肪的特点,富含维生素、矿物质、脂肪酸以及各种氨基酸。目前,我国的牛肉供应主要是以地方黄牛为主,如何提高地方黄牛的肌内脂肪含量从而改善肉品质,是我国畜牧行业的重大课题。前期研究表明,牛的饲养环境及营养水平可以对肌内脂肪含量产生影响,但遗传调控是影响肌内脂肪含量的决定性因素[1-3]。因此,从基因层面探究肌内脂肪沉积的调控机制意义重大。
ADIG基因是Kim等[4]为了研究脂肪细胞中新的分子通路而发现的新基因。研究表明,该基因是脂肪细胞分化过程中一个关键调节基因[5]。最近的研究指出,ADIG基因的异位表达可以增强牛肌卫星细胞的脂肪积累,并促进牛肌肉细胞向脂肪细胞转化[6-7],敲除ADIG基因会影响脂肪组织的生成[8]。因此,ADIG基因可能在脂肪形成过程中发挥重要功能。
目前,关于牛ADIG基因转录调控机制的研究尚不清楚,本试验以南阳牛为研究材料,选择可能与脂肪形成相关的ADIG基因为研究对象;构建牛ADIG基因组织表达谱;克隆获得牛ADIG基因启动子序列,确定其核心启动子区域;结合生物信息学预测调控牛ADIG基因的关键转录因子。为探究ADIG基因在肉牛分子水平上的改良培育提供理论依据。
1 材料与方法 1.1 试验材料待健康南阳公牛((24±2)月龄)屠宰后,迅速采集3头牛的心、肝、脾、肺、肾、肌肉和脂肪等7种组织,-80 ℃冰箱保存,同时采集血液样本-20 ℃保存备用。
1.2 牛不同组织ADIG基因mRNA表达量检测通过TRIzol法提取3头牛的肝、心、肺、脾、肌肉、肾及脂肪组织总RNA,质量检测合格后逆转录成cDNA。ADIG基因不同组织的相对表达量检测以cDNA为模板,设计定量引物(表 1)进行RT-PCR。反应体系:上、下游引物各0.5 μL,TB Green Premix Ex Taq Ⅱ (2×) 10 μL,cDNA 2 μL,ddH2O补至20 μL。反应条件:95 ℃预变性30 s;95 ℃5 s,60 ℃30 s,40个循环,试验设置3个生物学和技术学重复。
|
|
表 1 实时荧光定量引物信息 Table 1 Primers used in real-time PCR |
通过(https://www.ncbi.nlm.nih.gov/)NCBI数据库获得牛ADIG基因序列,结合Ensembl(https://asia.ensembl.org/index.html)网站,对牛ADIG基因启动子位置进行初步预测。MEGA 5.0软件分析牛ADIG基因的结构,构建ADIG蛋白质的进化树。
1.4 牛ADIG基因启动子克隆提取南阳牛血液基因组DNA,利用启动子全长引物ADIG-PF和ADIG-PR扩增牛ADIG基因的全长启动子(表 2)。体系为50 μL:2×PhantaⓇMax Master Mix 25 μL,50~100 ng基因组DNA,引物各2 μL,用ddH2O补齐。反应程序:95 ℃预变性30 s;95 ℃15 s,60 ℃20 s,72 ℃30 s,35个循环;72 ℃彻底延伸5 min,4 ℃10 min保存。PCR产物连接T载体,用于后续6条截短体扩增。
|
|
表 2 牛ADIG基因启动子及其缺失片段克隆引物 Table 2 Primers used in promoter and deletion fragment cloning of ADIG gene |
1.5.1 牛ADIG基因启动子逐段缺失片段的克隆 设计6条用于牛ADIG基因启动子不同截短片段的扩增引物ADIG-F1、ADIG-F2、ADIG-F3、ADIG-F4、ADIG-F5及ADIG-F6,1条固定下游引物ADIG-R0,引物分别添加Kpn Ⅰ和Bgl Ⅱ双酶切位点(表 2)。以牛ADIG基因启动子全长-2 186/+59片段为模板进行启动子逐段缺失片段扩增(反应体系和程序同“1.4”),测序鉴定后用于下一步试验。
1.5.2 牛ADIG基因启动子截短体载体构建 使用Kpn Ⅰ和Bgl Ⅱ在37 ℃下对pGL3-Basic载体和6个截短体进行双酶切,酶切产物连接成功后分别命名为:pADIG-1 621/+59、pADIG-1 288/+59、pADIG-850/+59、pADIG-589/+59、pADIG-321/+59和pADIG-79/+59,并用酶标仪进行纯度和浓度测定。
1.5.3 牛ADIG基因启动子核心区域鉴定 复苏293T细胞、牛原代脂肪细胞和3T3-L1细胞使用含有10%胎牛血清的DMEM培养基进行培养。将重组质粒和pRL-TK质粒以24∶1的比例分别共转染293T细胞、牛原代脂肪细胞以及3T3-L1细胞,根据荧光素酶活性检测试剂盒步骤,以pGL3-Basic为对照,分别检测出Firefly luciferase (F)和Renilla luciferase(R)的值,通过(F/R)的值确定牛ADIG基因启动子核心区域。
1.6 牛ADIG基因启动子关键转录因子预测通过(http://jaspar.genereg.net和http://gene-regulation.com/pu-b/programs/alibaba2/index.html)预测脂肪相关转录因子与牛ADIG基因核心启动子区域的结合位点,设置阈值大于90%,预测结果进行数据库对比并取交集,并对相关转录因子进行标注。
1.7 数据分析数据结果以“平均值±标准差”表示,结果使用SPSS17.0软件进行单因素方差分析。P < 0.05表示差异显著,用*表示;P < 0.01表示差异极显著,用**表示。
2 结果 2.1 牛不同组织ADIG基因的表达规律南阳牛ADIG基因的组织表达谱如图 1所示。ADIG基因在南阳牛肺(P < 0.05)、肾(P < 0.05)以及皮下脂肪(P < 0.01)组织中高表达。证明,ADIG基因在南阳牛脂肪组织中发挥重要作用。
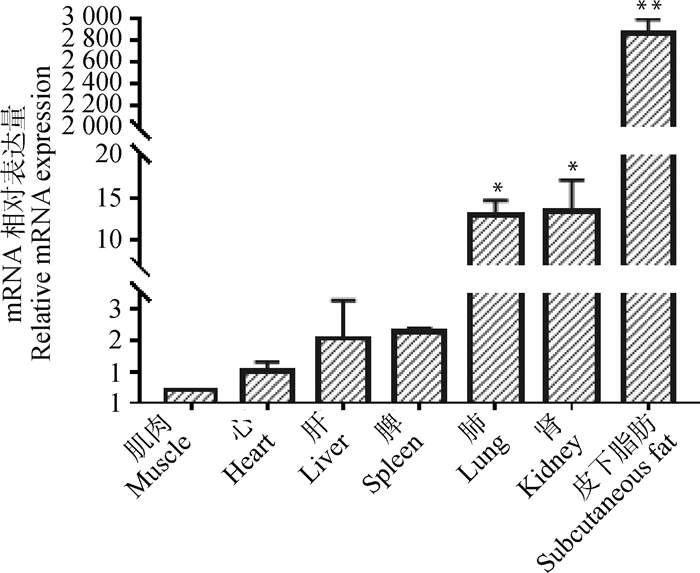
|
图 1 南阳牛不同组织中ADIG基因的相对表达情况 Fig. 1 Relative expression of ADIG genes in different tissues of Nanyang cattle |
牛ADIG基因位于13号染色体(NC_037340.1) 上,是一条含有3个外显子和2个内含子的全长为9 869 bp的序列,其蛋白序列由含有N末端和C末端的80个氨基酸构成,N端富含亮氨酸且具有核定位序列(PWSKRP),C末端富含酸性氨基酸,推测牛ADIG基因在调控过程中具有重要功能。以NCBI数据库公布的牛ADIG基因信息,确定其转录起始位点胞嘧啶(C)为+1位置,向上游查找2 245 bp的序列,预测其启动子区。不同ADIG蛋白质序列(山羊、马和猪)的进化树由MEGA 5.0软件构建。结果如图 2所示,表明ADIG蛋白在不同物种中同源性较高,尤其在牛和山羊间进化保守。

|
图 2 牛ADIG蛋白序列进化树 Fig. 2 Evolutionary tree of bovine ADIG protein sequence |
据牛ADIG的启动子序列信息,PCR扩增其启动子区(图 3A),并将PCR产物与T载体连接,测序证实其长度为2 245 bp且序列正确(图 3B)。
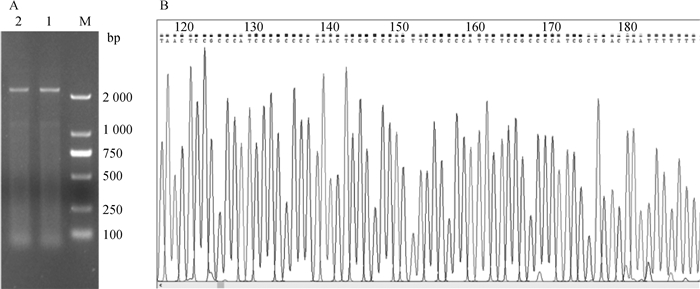
|
A.牛ADIG基因启动子PCR产物;B.ADIG基因pDM-18T载体测序部分图谱 A. Bovine ADIG gene promoter PCR product; B. Partial mapping of ADIG gene pDM-18T vector sequencing 图 3 牛ADIG基因启动子片段的PCR扩增 Fig. 3 PCR amplification of bovine ADIG gene promoter |
2.4.1 启动子不同截短体载体构建 使用ADIG-F1/ADIG-R0~ADIG-F6/ADIG-R0引物扩增获得牛ADIG基因启动子截短体,并连接pGL3-Basic载体,获得pADIG-1 621/+59、pADIG-1288/+59、pADIG-850/+59、pADIG-589/+59、pADIG-321/+59和pADIG-79/+59重组质粒。Kpn Ⅰ和Bgl Ⅱ双酶切鉴定结果如图 4所示,酶切后获得2条目的条带,其中包括4 818 bp空载体及1 680、1 347、909、648、380和138 bp的目的条带,条带长度与预期一致,重组质粒(OD260nm/OD280nm)在1.8~2.0之间,质量浓度全部在300 ng·μL-1以上,可用于下一步试验。
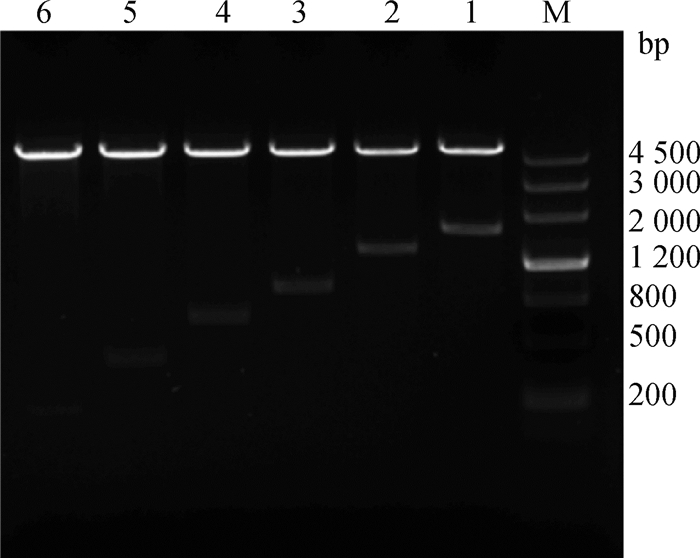
|
M.DNA相对分子质量标准;1~6.pADIG-1 621/+59、pADIG-1 288/+59、pADIG-850/+59、pADIG-589/+59、pADIG-321/+59和pADIG-79/+59经Kpn Ⅰ和Bgl Ⅱ双酶切结果 M. DL4500 DNA marker; 1-6. Double digestion results of Kpn Ⅰ and Bgl Ⅱ of pADIG-1 621/+59, pADIG-1 288/+59, pADIG-850/+59, pADIG-589/+59, pADIG-321/+59 and pADIG-79/+59 plasmids, respectively 图 4 牛ADIG基因启动子双荧光素报告重组质粒的双酶切鉴定 Fig. 4 Electrophoretic diagram of double digestion of dual-luciferase plasmid of bovine ADIG promoter |
2.4.2 牛ADIG基因启动子核心区域的确定 双荧光素酶报告基因缺失启动子片段的活性分析表明当牛ADIG基因启动子区-321/-79片段缺失时,双荧光素报告载体pADIG-79/+59在293T细胞(P < 0.05, 图 5A)、3T3-L1细胞(P < 0.01, 图 5B)和牛原代脂肪细胞(P < 0.05, 图 5C)中的活性较pADIG-321/+59都显著下降。表明,牛ADIG基因5′端上游2 245 bp序列是启动子区,具有调控基因转录活性功能,-321/-79 bp区域为牛ADIG基因启动子核心区。
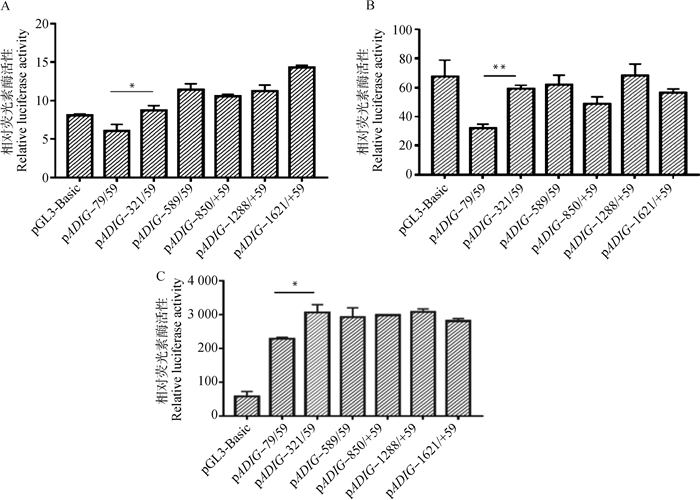
|
A.逐段缺失片段在293T细胞中的相对荧光活性;B.逐段缺失片段在3T3-L1细胞中的相对荧光活性;C. 逐段缺失片段在牛原代脂肪细胞中的相对荧光活性 A. Relative fluorescence activity of deletion fragment in 293T cells; B. Relative fluorescence activity of deletion fragment in 3T3-L1 cells; C.Relative fluorescence activity of deletion fragment in bovine primary adipocytes 图 5 牛ADIG基因核心启动子鉴定 Fig. 5 Identification of core promoter of ADIG gene |
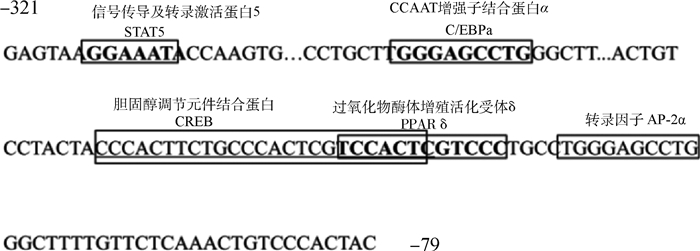
结合Genomatix和JASPAR等生物信息学手段预测牛ADIG基因启动子核心区域(-321/-79 bp) 关键转录因子的结合位点,结果如图 6所示,牛ADIG基因启动子核心区与CCAAT增强子结合蛋白α(C/EBPα)、过氧化物酶体增殖活化受体δ(PPARδ)、胆固醇调节元件结合蛋白(CREB)、信号传导及转录激活蛋白5(STAT5A)和AP-2α等相结合。表明C/EBPα、PPARδ、CREB、STAT5和AP-2α等对牛ADIG基因具有重要转录调节作用。
|
图 6 牛ADIG基因启动子核心区域转录因子预测 Fig. 6 Prediction of transcription factors in core region of bovine ADIG |
ADIG基因是3T3-L1细胞在诱导分化过程中筛选获得的一个新基因,且证明其只在分化成功后的脂肪细胞中表达[4]。对小鼠各组织定量发现,ADIG基因在脂肪组织中特异表达[5]。半定量的结果显示,ADIG基因在猪脂肪组织中高表达,且瘦肉型猪脂肪组织中的表达量远低于脂肪型猪[9]。敲除ADIG基因后会抑制小鼠脂肪形成[8]。本研究发现,ADIG基因在南阳牛肺(P < 0.05)、肾(P < 0.05)和皮下脂肪(P < 0.01)中高表达,且在不同物种进化过程中高度保守,ADIG基因在牛脂肪的沉积过程中可能发挥关键的调节作用。
本研究检测到牛ADIG基因-321/-79 bp为启动子核心区域,对其核心区域进行生物信息学分析,发现PPARδ、C/EBPα、CREB、STAT5和AP-2α等5个重要转录因子在该区域具有结合位点。转录因子对下游靶基因具有重要调控作用,从而对机体的生命活动产生影响[10]。C/EBPα是C/EBPS家族中的一员[11],还包括C/EBPβ、C/EBPγ、C/EBPδ、C/EBPε和C/EBPζ等4种亚型[12],该家族广泛参与细胞周期、个体的生长发育和体内免疫反应等基本生命活动[13-14]。C/EBPα具有p30kd和p42kd两种亚型[15],这两种亚型都参与各种重要生命活动[16]。C/EBPα在脂肪细胞的分化后期会显著性高表达,成脂标志基因也随之特异性高表达[17-19],异位表达C/EBPα可以促进成纤维细胞的脂肪积累[20-21],C/EBPα基因敲除小鼠出生后短时间内死亡,且体内脂肪含量远低于正常小鼠[22-23]。综上所述,C/EBPα对脂肪形成具有重要作用。本研究发现,牛ADIG基因启动子核心区有C/EBPα结合位点,所以推测,C/EBPα在牛ADIG基因的转录调节中发挥着关键的作用。
PPARδ广泛分布于全身的各个组织和器官,参与调节炎症、氧化应激、细胞增殖、细胞凋亡、凝血和其他生理学功能[24],激活PPARδ可以提高胰岛素敏感性和调节机体能量代谢[25]。脂肪组织中PPARδ可以作为脂肪酸感受器调节脂肪酸氧化和线粒体呼吸等生命活动的基因表达,从而预防肥胖发生[26-27],还有研究发现,被前列环素及维甲酸激活的PPARδ具有调控基因转录的功能[28]。CREB通过与靶基因的cAMP相结合发挥转录调控作用[29]。CREB在细胞的各种生理学过程中起关键作用,并参与癌症和其他疾病的调节[30]。研究表明,CREB通过磷酸化激活PPARγ、C/EBPα和C/EBPβ在脂肪细胞分化早期发挥作用[31],也通过调节多种蛋白和激酶对脂肪细胞分化后期产生影响[32]。
STAT5、STAT1、STAT2、STAT3、STAT4和STAT6全部属于STAT家族[33],在转录激活与信号转导中起关键作用[34]。STAT5是葡萄糖和脂肪代谢路径中一个重要的调节因子[35-37]。研究表明,STAT5随脂肪细胞的分化稳定表达,干扰STAT5后显著降低脂肪细胞分化标志基因的表达[38]。作为AP-2转录因子家族中最早期鉴定的转录因子,AP-2α在细胞增殖、胚胎发育,分化和凋亡、软骨形成以及癌症等生理疾病调控中发挥重要作用[39-40]。研究表明,AP-2α在3T3-L1前脂肪细胞中参与电子传递、脂肪酸代谢以及协同调控脂肪细胞分化[41],AP-2α通过负调控C/EBPα抑制脂联素表达[42-43]。综上,C/EBPα、PPARδ、CREB、STAT5和AP-2α在个体各种疾病及生命活动和脂肪沉积过程中发挥重要的调节作用。
4 结论本研究表明,牛ADIG基因分别在肺、肾以及皮下脂肪组织中均有表达且在皮下脂肪中极显著高表达;其核心区域在-321/-79 bp;结合生物信息学初步鉴定C/EBPα、PPARδ、CREB、STAT5和AP-2α转录因子对ADIG基因的转录活性有重要的调控作用。以上结果为探究ADIG基因在肉牛分子水平上的改良培育提供了理论依据。
| [1] |
CECCHINATO A, DE MARCHI M, PENASA M, et al. Genetic analysis of beef fatty acid composition predicted by near-infrared spectroscopy[J]. J Anim Sci, 2012, 90(2): 429-438. DOI:10.2527/jas.2011-4150 |
| [2] |
ENGLISHBY T M, BANOS G, MOORE K L, et al. Genetic analysis of carcass traits in beef cattle using random regression models1[J]. J Anim Sci, 2016, 94(4): 1354-1364. DOI:10.2527/jas.2015-0246 |
| [3] |
WANG Y N, ZHANG F, MUKⅡBI R, et al. Genetic architecture of quantitative traits in beef cattle revealed by genome wide association studies of imputed whole genome sequence variants: Ⅱ: carcass merit traits[J]. BMC Genomics, 2020, 21(1): 38. DOI:10.1186/s12864-019-6273-1 |
| [4] |
KIM J Y, TILLISON K, SMAS C M. Cloning, expression, and differentiation-dependent regulation of SMAF1 in adipogenesis[J]. Biochem Biophys Res Commun, 2004, 326(1): 36-44. DOI:10.1016/j.bbrc.2004.10.200 |
| [5] |
HONG Y H, HISHIKAWA D, MIYAHARA H, et al. Up-regulation of adipogenin, an adipocyte plasma transmembrane protein, during adipogenesis[J]. Mol Cell Biochem, 2005, 276(1-2): 133-141. DOI:10.1007/s11010-005-3673-0 |
| [6] |
REN G, ESKANDARI P, WANG S Q, et al. Expression, regulation and functional assessment of the 80 amino acid Small Adipocyte Factor 1 (Smaf1) protein in adipocytes[J]. Arch Biochem Biophys, 2016, 590: 27-36. DOI:10.1016/j.abb.2015.09.019 |
| [7] |
LIU Y, JIANG B J, FU C Z, et al. Cloning and characterization of adipogenin and its overexpression enhances fat accumulation of bovine myosatellite cells[J]. Gene, 2017, 601: 27-35. DOI:10.1016/j.gene.2016.11.040 |
| [8] |
ALVAREZ-GUAITA A, PATEL S, LIM K, et al. Phenotypic characterization of Adig null mice suggests roles for adipogenin in the regulation of fat mass accrual and leptin secretion[J]. Cell Rep, 2021, 34(10): 108810. DOI:10.1016/j.celrep.2021.108810 |
| [9] |
杨红文, 郑嵘, 李凤娥, 等. 猪小脂肪细胞因子1(SMAF1)基因cDNA的克隆及表达谱分析[J]. 农业生物技术学报, 2007, 15(1): 11-14. YANG H W, ZHENG R, LI F E, et al. Cloning and expression analysis of porcine small adipocyte Factor 1 (SMAF1) gene[J]. Journal of Agricultural Biotechnology, 2007, 15(1): 11-14. DOI:10.3969/j.issn.1674-7968.2007.01.003 (in Chinese) |
| [10] |
LAMBERT S A, JOLMA A, CAMPITELLI L F, et al. The human transcription factors[J]. Cell, 2018, 172(4): 650-665. DOI:10.1016/j.cell.2018.01.029 |
| [11] |
TSUKADA J, YOSHIDA Y, KOMINATO Y, et al. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation[J]. Cytokine, 2011, 54(1): 6-19. DOI:10.1016/j.cyto.2010.12.019 |
| [12] |
CAO Z, UMEK R M, MCKNIGHT S L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells[J]. Genes Dev, 1991, 5(9): 1538-1552. DOI:10.1101/gad.5.9.1538 |
| [13] |
WANG Y G, DASSO M. SUMOylation and deSUMOylation at a glance[J]. J Cell Sci, 2009, 122(Pt 23): 4249-4252. |
| [14] |
CORRELL K A, EDEEN K E, REDENTE E F, et al. TGF beta inhibits HGF, FGF7, and FGF10 expression in normal and IPF lung fibroblasts[J]. Physiol Rep, 2018, 6(16): e13794. DOI:10.14814/phy2.13794 |
| [15] |
ANIRUDH S, ROSENBERGER A, SCHWARZENBERGER E, et al. TNFα rescues dendritic cell development in hematopoietic stem and progenitor cells lacking C/EBPα[J]. Cells, 2020, 9(5): 1223. DOI:10.3390/cells9051223 |
| [16] |
CHEN L F, YUAN R F, WEN C Y, et al. E3 ubiquitin ligase UBR5 promotes pancreatic cancer growth and aerobic glycolysis by downregulating FBP1 via destabilization of C/EBPα[J]. Oncogene, 2021, 40(2): 262-276. DOI:10.1038/s41388-020-01527-1 |
| [17] |
ZAIDI N, LUPIEN L, KUEMMERLE N B, et al. Lipogenesis and lipolysis: the pathways exploited by the cancer cells to acquire fatty acids[J]. Prog Lipid Res, 2013, 52(4): 585-589. DOI:10.1016/j.plipres.2013.08.005 |
| [18] |
WU J, BOSTRÖM P, SPARKS L M, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human[J]. Cell, 2012, 150(2): 366-376. DOI:10.1016/j.cell.2012.05.016 |
| [19] |
ZABORSKA K E, WAREING M, EDWARDS G, et al. Loss of anti-contractile effect of perivascular adipose tissue in offspring of obese rats[J]. Int J Obes, 2016, 40(8): 1205-1214. DOI:10.1038/ijo.2016.62 |
| [20] |
MORIGNY P, HOUSSIER M, MOUISEL E, et al. Adipocyte lipolysis and insulin resistance[J]. Biochimie, 2016, 125: 259-266. DOI:10.1016/j.biochi.2015.10.024 |
| [21] |
WEISZENSTEIN M, MUSUTOVA M, PLIHALOVA A, et al. Adipogenesis, lipogenesis and lipolysis is stimulated by mild but not severe hypoxia in 3T3-L1 cells[J]. Biochem Biophys Res Commun, 2016, 478(2): 727-732. DOI:10.1016/j.bbrc.2016.08.015 |
| [22] |
CHEN W B, ZHANG L X, ZHAO Y K, et al. C/EBPα-mediated transcriptional activation of miR-134-5p entails KPNA3 inhibition and modulates focal hypoxic-ischemic brain damage in neonatal rats[J]. Brain Res Bull, 2020, 164: 350-360. DOI:10.1016/j.brainresbull.2020.08.006 |
| [23] |
ZHU Y, CHEN X Q, MI L L, et al. Sumoylation of CCAAT-enhancer-binding protein α inhibits lung differentiation in Bronchopulmonary dysplasia model rats[J]. J Cell Mol Med, 2020, 24(12): 7067-7071. DOI:10.1111/jcmm.15310 |
| [24] |
DOKTOROVA M, ZWARTS I, ZUTPHEN T V, et al. Intestinal PPARδ protects against diet-induced obesity, insulin resistance and dyslipidemia[J]. Sci Rep, 2017, 7(1): 846. DOI:10.1038/s41598-017-00889-z |
| [25] |
VÁZQUEZ-CARRERA M. Unraveling the effects of PPARβ/δ on insulin resistance and cardiovascular disease[J]. Trends Endocrinol Metab, 2016, 27(5): 319-334. DOI:10.1016/j.tem.2016.02.008 |
| [26] |
BRUNMEIR R, XU F. Functional regulation of PPARs through post-translational modifications[J]. Int J Mol Sci, 2018, 19(6): 1738. DOI:10.3390/ijms19061738 |
| [27] |
LIU Y, COLBY J K, ZUO X S, et al. The role of PPAR-δ in metabolism, inflammation, and cancer: many characters of a critical transcription factor[J]. Int J Mol Sci, 2018, 19(11): 3339. DOI:10.3390/ijms19113339 |
| [28] |
MAGADUM A, ENGEL F. PPARβ/δ: linking metabolism to regeneration[J]. Int J Mol Sci, 2018, 19(7): 2013. DOI:10.3390/ijms19072013 |
| [29] |
LEE B, LI A Q, HANSEN K F, et al. CREB Influences Timing and Entrainment of the SCN Circadian Clock[J]. J Biol Rhythms, 2010, 25(6): 410-420. DOI:10.1177/0748730410381229 |
| [30] |
MACHADO G D B, DE FREITAS B S, FLORIAN L Z, et al. G protein-coupled oestrogen receptor stimulation ameliorates iron-and ovariectomy-induced memory impairments through the cAMP/PKA/CREB signalling pathway[J]. J Neuroendocrinol, 2019, 31(10): e12780. |
| [31] |
BARTOLOTTI N, LAZAROV O. CREB signals as PBMC-based biomarkers of cognitive dysfunction: A novel perspective of the brain-immune axis[J]. Brain Behav Immun, 2019, 78: 9-20. DOI:10.1016/j.bbi.2019.01.004 |
| [32] |
CHEN X, DAI J C, GREENFIELD E M. Termination of immediate-early gene expression after stimulation by parathyroid hormone or isoproterenol[J]. Am J Physiol Cell Physiol, 2002, 283(5): C1432-C1440. DOI:10.1152/ajpcell.00221.2002 |
| [33] |
DARNELL J E Jr. STATs and gene regulation[J]. Science, 1997, 277(5332): 1630-1635. DOI:10.1126/science.277.5332.1630 |
| [34] |
FURIGO I C, RAMOS-LOBO A M, FRAZÃO R, et al. Brain STAT5 signaling and behavioral control[J]. Mol Cell Endocrinol, 2016, 438: 70-76. DOI:10.1016/j.mce.2016.04.019 |
| [35] |
ZHANG Y Z, ZHOU L T, ZHANG Z M, et al. Effects of Di (2-ethylhexyl) phthalate and high-fat diet on lipid metabolism in rats by JAK2/STAT5[J]. Environ Sci Pollut Res, 2020, 27(4): 3837-3848. DOI:10.1007/s11356-019-06599-5 |
| [36] |
NAKAMURA M T, YUDELL B E, LOOR J J. Regulation of energy metabolism by long-chain fatty acids[J]. Prog Lipid Res, 2014, 53: 124-144. DOI:10.1016/j.plipres.2013.12.001 |
| [37] |
FURIGO I C, MELO H M, SILVA N M L E, et al. Brain STAT5 signaling modulates learning and memory formation[J]. Brain Struct Funct, 2018, 223(5): 2229-2241. DOI:10.1007/s00429-018-1627-z |
| [38] |
张玉超. 生长激素与其信号转导分子STAT5B对脂肪细胞的影响及其与胰岛素信号的Crosstalk[D]. 济南: 山东大学, 2013. ZHANG Y C. The impact of Growth hormone and STAT5B signaling on adipocyte differentiation and the associated crosstalk with insulin[D]. Ji'nan: Shandong University, 2013. (in Chinese) |
| [39] |
GARCIA M F, MOORE C D, SCHULZ K N, et al. Structural features of transcription factors associating with nucleosome binding[J]. Mol Cell, 2019, 75(5): 921-932.e6. DOI:10.1016/j.molcel.2019.06.009 |
| [40] |
KOŁAT D, KAŁUZIŃSKA Ż, BEDNARE A K, et al. The biological characteristics of transcription factors AP-2α and AP-2γ and their importance in various types of cancers[J]. Biosci Rep, 2019, 39(3): BSR20181928. DOI:10.1042/BSR20181928 |
| [41] |
周求知, 王义军, 刘庆文, 等. 小鼠前脂肪细胞中AP-2α相互作用蛋白质的分离和鉴定[J]. 激光生物学报, 2011, 20(2): 199-202. ZHOU Q Z, WANG Y J, LIU Q W, et al. Separation and identification of AP-2α Interacting proteins from mice pre-adipocyte cells[J]. Acta Laser Biology Sinica, 2011, 20(2): 199-202. DOI:10.3969/j.issn.1007-7146.2011.02.012 (in Chinese) |
| [42] |
JIANG M S, TANG Q Q, MCLENITHAN J, et al. Derepression of the C/EBPα gene during adipogenesis: identification of AP-2α as a repressor[J]. Proc Natl Acad Sci U S A, 1998, 95(7): 3467-3471. DOI:10.1073/pnas.95.7.3467 |
| [43] |
李文. MiR-122在3T3-L1脂肪前体细胞诱导分化成熟过程中调控机制的研究[D]. 长沙: 湖南师范大学, 2012. LI W. The regulatory mechanism study of MIR-122on preadipocytes 3T3-L1 differentiation and maturation[D]. Changsha: Hunan Normal University, 2012. (in Chinese) |
(编辑 郭云雁)



