染色质组装因子1 (chromatin assembly factor-1,CAF-1),是由p150、p60、p48三个亚单位按1:1:1组合构成的三聚体组蛋白伴侣[1-2]。CAF-1主要功能是在DNA复制中与增殖细胞核抗原(proliferating cell nuclear antigen, PCNA)相互作用,负责募集组蛋白H3、H4沉积在新合成的DNA上以促进核小体装配。CAF-1不仅参与DNA复制和修复后染色质装配,而且参与细胞增殖调控、胚胎干细胞的表观遗传学变化及异染色质的合成和修复。
随着细胞分化,染色质越来越凝集。体细胞重编程为全能性的胚胎,首先要进行染色质的去凝集。在卵母细胞介导的重编程过程中,组蛋白发生置换,并擦除来自体细胞的抑制型表观修饰使染色质去凝聚。但是,体细胞克隆胚胎重编程过程中,来自体细胞的遗留表观遗传修饰擦除不完全,抑制胚胎基因组激活,导致胚胎发育失败、克隆动物死亡。而CAF-1通过H3K9 me3等组蛋白修饰在异染色质形成、染色质凝集事件上扮演着重要角色。因此,本文从CAF-1的结构、生物学功能和参与体细胞重编程的研究3部分作了简要的综述。
1 CAF-1的结构CAF-1包含3个亚基并依其大小命名:分别为150 ku亚基(p150亚基,CHAF1a)、60 ku亚基(p60,CHAF1b)和48 ku亚基(p48,RbAp48)。CAF-1复合体在不同物种上都参与DNA的合成和修复、染色质的组装,具有高度的结构和功能保守性。
CHAF1a的N端存在一个在体外具有强活性的PIP (PCNA相互作用肽),以及一个小泛素样修饰相互作用区域,这是与SUMO2/3相互作用所必需的[3]。在PxVxL区域还存在一个HP1(异染色质蛋白1)相互作用域[4]。一个PEST结构域驱动蛋白质的快速降解。在PEST结构域之后是一个KER结构域,这是一个高酸性的区域,被认为有利于与组蛋白的相互作用。在KER域内是另一个PIP,已被试验证实其在体内与PCNA相互作用和核小体形成有关。随后,还存在一个与CHAF1b相互作用域和ED区域,ED区域被认为与组蛋白和有翼螺旋结构域(WHD)结合[5]。一个与p48相互作用区域和一个有翼螺旋结构域位于CHAF1a的C端(图 1)[6]。
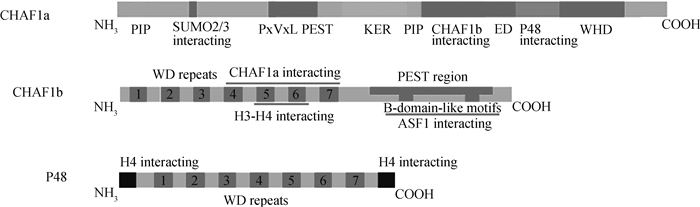
|
图 1 CAF-1复合体的结构(根据参考文献[14-15]修改) Fig. 1 Architecture of the CAF-1 complex (modified from reference [14-15]) |
CHAF1b位于21号染色体上,是由位于N端7×WD重复区域、位于C端B区结构域和PEST结构域3部分组成(图 1)。CHAF1b的7×WD重复区域过去被认为是提供CHAF1b-/ASF1a/H3/H4复合物作用所需的位点,研究证明,CHAF1b的7×WD重复区域并不是结合ASF1a/b的区域[7]。研究表明,CHAF1b依赖于B区结构域与ASF1a结合,形成CHAF1b/ASF1a/H3/H4复合物,再通过与CHAF1a和PCNA相互作用,定位在复制叉位置募集组蛋白,完成DNA的复制和核小体的组装。而这种结合是CHAF1b的B区结构形成β-发夹能与ASF1a氨基端核心区域的β-sandwich结构产生特异性的交互反应[8]。此外,CHAF1b的B区结构域与HIRA同源,因此,ASF1a与HIRA或CHAF1b的结合是相互排斥的,这表明HIRA表达可能是调节CHAF1b活性的一种间接方法[9]。CHAF1b的PEST结构域也发挥重要的作用。shRNA介导的CHAF1a的敲除会导致CHAF1b的表达减少则证明这一点[10]。
p48是7×WD重复区域,还包括两个分别在N和C末端的α-helical区域,它们可促进与H4组蛋白的结合(图 1)。它可能与组蛋白去乙酰化酶HDAC1的催化亚基密切相关,这表明它可能有助于新合成的H4在核小体中沉积后去乙酰化[11-13]。p48也是其他几个染色质调节复合物的组成部分,如核心蛋白复合体PRC2 (PRC2)、核小体重塑因子(NURF)、核小体重塑和去乙酰酶(NURD) 和组蛋白去乙酰化酶(HDAC1),提示该亚基在组蛋白修饰酶与其底物之间起着分子桥梁作用。除了与核心蛋白复合体PRC2作用外,p48也被认为在NURD重塑/去乙酰化酶复合物和NURF重塑复合物中发挥重要作用。
2 CAF-1的生物学功能 2.1 CAF-1参与DNA复制偶联的核小体组装在真核生物DNA复制过程中,CAF-1作为组蛋白伴侣,将组蛋白H3-H4异二聚体沉积到新合成的DNA上,促进复制叉后的核小体组装(图 2)[16-17]。在这个过程中组蛋白H3-H4二聚体与乙酰化的H3K56残基首先被组蛋白伴侣ASF1捕获,然后转移到CAF-1,这一过程是通过ASF1与CHAF1b直接相互作用介导的[16, 18]。据报道,组蛋白H3K56残基在人体内被CBP或Gcn5乙酰化,在酵母体内被Rtt109乙酰化[19-22]。乙酰化后的H3K56增加了酵母模型中CAF-1与H3-H4结合的亲和力,促进了CAF-1介导的核小体组装[19-23]。据报道,酵母和人类细胞中的HAT1负责H4K5和H4K12的乙酰化。然而,与H3K56的乙酰化作用不同,H4K5或K12的乙酰化作用降低了哺乳动物细胞中H3-H4与CAF-1的结合,而H3K56的乙酰化作用增强了CAF-1与H3-H4的体外结合亲和力[16, 19]。最后,CAF-1通过与“滑动的夹子”PCNA相互作用,将组蛋白H3-H4四聚体招募到复制叉,完成染色质组装。
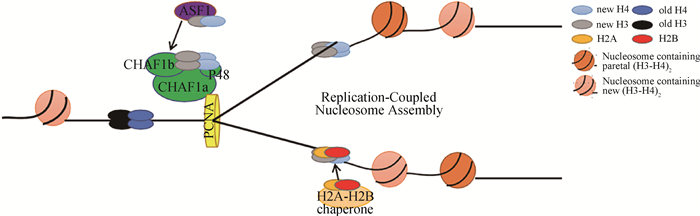
|
图 2 CAF-1参与DNA复制偶联型核小体组装过程(根据参考文献[4]修改) Fig. 2 CAF-1 participates in the DNA replication-coupled nucleosome assembly process (modified from reference [4]) |
CAF-1主要功能就是在S期,与ASF1a和/H3/H4组蛋白结合成CHAF1b/ASF1a/H3/H4复合物再与PCNA相互作用,定位在复制叉位置募集组蛋白,完成DNA复制耦联的核小体组装。因此,CHAF1b对S期过程维持是至关重要的。比如,敲除CHAF1b将使动物细胞由于染色质组装缺陷而停滞在S期,导致细胞死亡。敲降CHAF1b的HUH-7细胞导致细胞周期G0/G1期细胞比例减少,而S期细胞数目增多,G2/M期细胞数量变化不大,表明敲减CHAF1b基因减少G1期细胞的数量并增加S期细胞比例,导致细胞在不同阶段的分布发生变化[24]。干扰CHAF1b的表达可导致95-D细胞的细胞周期停滞。G1期细胞百分比显著增加,而S期细胞的比例减少,说明CHAF1b促进了细胞周期S期的DNA复制和核小体组装[25]。用CAF-1的特异性抑制剂HA-p150C,破坏内源性CHAF1a和CHAF1b之间的相互作用,阻止CHAF1b与染色质和DNA合成位点紧密结合,导致S期停滞。
2.3 CAF-1在细胞增殖中的功能CAF-1的主要功能是S期将新合成的H3/H4二聚体递送至复制叉的位置。在分裂间期局限在核仁,在S期集中于核内DNA复制的位点[26],可作为区别增殖期细胞和静止期细胞的标志物。因此,CHAF1b是和细胞增殖呈正相关的一种组蛋白伴侣[27]。CAF-1参与复制偶联染色质组装过程,其中CAF-1的消耗导致Chk1的激活和S期阻滞。沉默CHAF1b基因能抑制肝癌细胞的增殖能力,抵抗细胞死亡[28]。然而,在小鼠ES细胞中,CHAF1a敲降后DNA复制仍然进行。大量研究证明,CAF-1能通过调整S期特定染色质重组来促进细胞周期的进程从而影响细胞的增殖[29]。
2.4 CAF-1在细胞分化中的功能细胞分化是细胞类型特化的过程,在细胞发育过程中受特定基因表达的调控。研究表明,CHAF1a的下调足以通过神经母细胞瘤基因的失调和H3K9 me3修饰的整体减少导致代谢基因表达的缺失而诱导神经母细胞瘤细胞分化[30]。另一项研究表明,在MLL-AF9白血病细胞中,CHAF1b通过染色质中离散位点的直接积累来维持其未分化状态[28]。然而,在纤维母细胞中,CHAF1b也可以控制染色质可及性来抑制干细胞基因的表达,从而保持体细胞的分化状态[31]。因此,CHAF1b对基因具有细胞类型特异性作用调节: 在分化细胞中,CHAF1b保持细胞同一性;在恶性细胞中,CHAF1b保持未分化状态。这种CHAF1b的差异活性可能是由细胞核内的系特异性转录因子和染色质可达区域的组成介导的[28]。
2.5 CAF-1参与组蛋白修饰CAF-1一方面直接识别表观遗传。CAF-1将组蛋白H3-H4复合物靶向到DNA上。首先是新合成组蛋白H3和H4上面位点的乙酰化,然后CAF-1识别H4K5、H4K12和H3K56的乙酰化[32]。CAF-1另一方面间接调节表观遗传。在果蝇上研究发现,CAF-1对异染色质区域组蛋白H3K9位点的甲基化水平以及HP1蛋白的募集具有调节作用,从而决定异染色质结构的形成与稳定性的维持[33]。实际上,CHAF1a和HP1a互作从而维持果蝇异染色质[34]。小鼠胚胎干细胞的研究中,CHAF1a的缺失导致了包括HP1和H3K9 me3在内的构成性异染色质特征的严重改变,而中心性异染色质H3K9 me3和H4K20 me3的表观遗传标记完全被消除。甲基化CpG结合蛋白MBD1募集组蛋白甲基化酶SETDB1到CHAF1a形成MBD1/ SETDB1/CAF-1复合体,促使H3K9位点甲基化,对异染色质的形成具有重要作用。最近研究显示,CAF-1联合Kdm1a、Hdac1/2参与调节异染色质ERVs的转录抑制[35-36]。Hatanaka等[37]详细研究了CHAF1a通过调节组蛋白H3.1/3.2与H3.3的置换将H3K9 me3、H4K20 me3等多种抑制型组蛋白修饰富集到对应区域。在植物中,CAF-1与H3K27甲基转移酶, 即PRC2相互作用; 有助于在DNA复制过程中保护H3K27 me3介导的沉默染色质[38]。与此观点一致,在小鼠胚胎干细胞中CAF-1也与PRC2复合物相互作用(图 3)[39]。
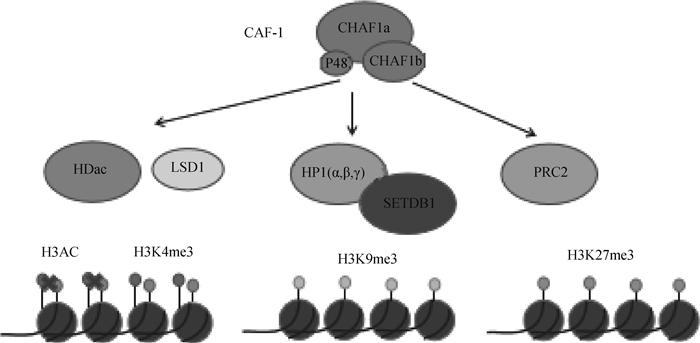
|
图 3 CAF-1参与组蛋白修饰(根据参考文献[40]修改) Fig. 3 CAF-1 is involved in histone modification (modified from reference [40]) |
体细胞注入卵母细胞后,在卵胞质中重编程因子的作用下,将体细胞重编程为全能性的胚胎[41]。在此过程中,染色质重编程是克隆胚胎发育成败的关键所在,主要包括染色质去凝集、染色质重构和组蛋白修饰等。但是,由于来自体细胞的遗留表观遗传修饰擦除不完全以及抑制型组蛋白的异常嵌入,将抑制胚胎基因组激活,导致胚胎发育失败以及克隆动物死亡[42-44]。
3.1 CAF-1在早期胚胎发育中的作用在早期胚胎发育过程中,CHAF1b蛋白表达水平在2细胞期胚胎之后逐渐升高,这可能主要与在发育过程中细胞复制的高需求有关。在小鼠上,完全定点突变CHAF1a使小鼠胚胎发育阻滞在16-细胞期胚胎,表明CHAF1a是早期胚胎和多能性干细胞的异染色质组装和维持所必需的[45]。敲降CHAF1a,降低组蛋白H3.1水平,升高H3.3水平,阻抑异染色质形成,可使小鼠胚胎的发育阻滞在囊胚前[46]。Hatanaka等[37]详细研究了CHAF1a在小鼠附植前胚胎上的作用机理, CHAF1a通过调节组蛋白H3.1/3.2与H3.3的置换将H3K9 me3、H4K20 me3等多种抑制型组蛋白修饰富集到异染色质区LINE1等反转座子,从而维持反转座子等异染色质区域的稳定。
3.2 CAF-1是体细胞重编程的重要障碍CAF-1是动物乃至植物上细胞分化过程中的关键蛋白。Cheloufi等[31]通过大范围RNAi筛选技术发现,CAF-1两个亚基CHAF1a和CHAF1b是体细胞重编程障碍因子。通过敲降CHAF1a或CHAF1b可显著提高iPS重编程效率。抑制CAF-1表达除了可促进分化细胞诱导为多能性细胞,还促使细胞转分化,说明CAF-1守卫体细胞特性,阻抑转录因子诱导细胞命运的转换。但CAF-1在体细胞克隆胚胎重编程过程中的作用及其机理尚不清楚。在小鼠胚胎干细胞上敲降CHAF1b,可使多能性的ES细胞转化为更多的全能性ES细胞,这种全能性ES细胞用于核移植后重编程效率显著提高, 同时促进了类似于两细胞阶段卵裂球的全能样细胞的出现[47]。表明CAF-1是体细胞重编程的重要障碍。
在iPSCs的重编程过程中,CAF-1的抑制作用于局部的增强子元件,使它们更容易与转录因子结合。在小鼠和人早期核移植胚胎上,位于异染色质区的反转录转座子重复序列(LINE和LTR等)遗留着来自体细胞的抑制型组蛋白修饰H3K9 me3,使得这些区域沉默,被称为抗重编程区(reprogramming-resistant regions, RRRs)[45, 48-49]。而这些异染色质区重复序列在小鼠附植前胚胎高表达,对胚胎基因组激活和胚胎发育具有重要作用[50-52]。研究人员通过擦除H3K9 me3,重新激活RRRs,显著提高小鼠克隆效率并促进人核移植胚胎发育[45, 48]。当然,这些异染色质区重复序列通常在受精卵来源的胚胎中2细胞期胚胎活跃,但在SCNT胚胎中仍然沉默,从而阻碍了克隆小鼠出生。相比ES细胞,敲降CHAF1b获得的全能性ES细胞用于核移植,小鼠胚胎的RRRs区域大量激活(图 4)[35]。
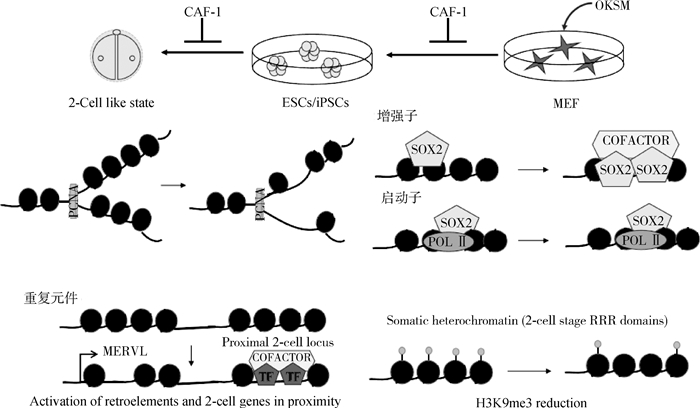
|
图 4 CAF-1是体细胞重编程的重要障碍(根据参考文献[40]修改) Fig. 4 CAF-1 is an important obstacle to somatic cell reprogramming (modified from reference[40]) |
在通过CAF-1沉默将ESC转换为类似两细胞阶段的全能样细胞的状态期间,染色质可及性减小,从而导致内源性逆转录元件(例如MERVL)和邻近基因的激活。MERVL是小鼠2-细胞期胚胎特异性表达的反转座子,MERVL的转录可以激活2-细胞期特异基因的表达,对于胚胎正常发育至关重要。全基因组siRNA筛发现,CAF-1是主要的MERVL抑制因子[35],在小鼠2-细胞期胚胎上CHAF1b表达水平和MERVL表达负相关,MERVL转录的位置检测不到CHAF1b,敲降CHAF1b可上调MERVL的表达[35]。与此同时证明敲除CAF-1导致内源性逆转录病毒元件(例如MERVL)高表达(图 4)[45]。
3.3 组蛋白H3.1的异常嵌入可能导致克隆胚胎重编程失败在小鼠核移植中,供体细胞来源的H3.2、H3.3和H2A迅速地被卵胞质中的组蛋白H3变体和H2AX替换。克隆胚胎上组蛋白H3.2和3.3与体外受精胚胎一样,而组蛋白变体H3.1嵌入到克隆胚胎但未嵌入体外受精胚胎[53]。组蛋白H3.1上富集抑制型组蛋白修饰,与转录抑制相关;而组蛋白H3.3主要富集在转录起始位点、增强子以及转录激活的基因上,和转录激活相关。多项研究显示,母源组蛋白H3.3是重要的重编程因子,体细胞核移植到卵胞质后,组蛋白H3.3和体细胞组蛋白发生置换,并启动克隆胚胎基因组激活,在克隆胚胎重编程过程中扮演重要角色[54]。所以,组蛋白变体H3.1异常嵌入到克隆胚胎可能是克隆胚胎重编程失败的重要原因。
组蛋白H3.1上富集的抑制型组蛋白修饰H3K9 me3是染色质凝集状态的标志。H3K9 me3形成异染色质蛋白HP1结合位点,HP1识别并结合在甲基化修饰的组蛋白,募集DNA甲基化转移酶DNMT,使该位点DNA甲基化修饰[55]。另外,HP1结合甲基化的组蛋白后可以招募组蛋白甲基化转移酶Suv39 h1,使附近的组蛋白也发生组蛋白甲基化,向两边延伸,形成异染色质,使染色质发生凝集。
3.4 CAF-1介导H3.1/3.2与H3.3的置换可能导致克隆胚胎重编程失败CAF-1是组蛋白H3.1的伴侣蛋白,促进H3.1嵌入到新合成的DNA [56]。组蛋白H3.1上富集抑制型组蛋白修饰,与转录抑制相关[57];而组蛋白H3.3主要富集在转录起始位点、增强子以及转录激活的基因上,和转录激活相关[58]。由于CAF-1是组蛋白变体H3.1的伴侣蛋白,在小鼠早期胚胎负责组蛋白3.1和组蛋白3.3置换,置换过程中将抑制型表观修饰例如H3K9 me3富集到对应区域[37]。敲降CHAF1a导致组蛋白H3.1去除,而促进H3.3的嵌入,阻抑异染色质形成[46, 59]。同样,敲降CHAF1b调控组蛋白H3.1嵌入到新合成的DNA上,而促进组蛋白H3.3沉积[60]。
研究显示,CAF-1和异染色质蛋白HP1以及H3K9 me3的互作,能够促进细胞分化过程中异染色质形成和染色质的凝集。在诱导多能性干细胞(iPS)的研究中发现,CAF-1是体细胞重编程的抑制因子,敲降其表达可促进体细胞向iPS的转化,显著提高iPS重编程效率[61-62]。本课题组研究发现,敲降CHAF1b可上调牛克隆胚胎组蛋白变体H3.3的含量,降低组蛋白H3K9 me3丰度,并显著提高牛克隆胚胎的桑椹胚和囊胚发育率(未发表数据)。推断在牛克隆胚胎上敲降CHAF1b很可能引起组蛋白变体置换,并在置换过程中将高丰度的抑制型组蛋白修饰例如H3K9 me3去除,导致原有被其抑制的抗重编程区重新激活,从而促进牛克隆胚胎发育。
4 小结自从1986年第1次报道CAF-1,距今已经有34年。在这些年的研究中,CAF-1的结构已清晰,正如前文所述,CAF-1由p150、p60、p48三个亚基的组蛋白伴侣,每个亚基的不同结构域都行使着不同的功能,它们负责与不同的蛋白相互作用。当然,CAF-1也参与多种生物学进程,包括DNA复制偶联的核小体组装、细胞周期、细胞增殖和细胞分化等。最近几年来,CAF-1被不断证明是体细胞重编程障碍因子。敲降CAF-1可显著提高iPS重编程效率。抑制CAF-1表达除了可促进分化细胞诱导为多能性细胞,还促使细胞转分化,说明CAF-1守卫体细胞特性,阻抑转录因子诱导细胞命运的转换。结合本课题组前期的试验结果,猜测CAF-1介导H3.1/3.2与H3.3的置换可能会导致克隆胚胎重编程失败。
| [1] | STILLMAN B. Chromatin assembly during SV40 DNA replication in vitro[J]. Cell, 1986, 45(4): 555–565. DOI: 10.1016/0092-8674(86)90287-4 |
| [2] | SMITH S, STILLMAN B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro[J]. Cell, 1989, 58(1): 15–25. DOI: 10.1016/0092-8674(89)90398-X |
| [3] | UWADA J, TANAKA N, YAMAGUCHI Y, et al. The p150 subunit of CAF-1 causes association of SUMO2/3 with the DNA replication foci[J]. Biochem Biophys Res Commun, 2010, 391(1): 407–413. DOI: 10.1016/j.bbrc.2009.11.071 |
| [4] | THIRU A, NIETLISPACH D, MOTT H R, et al. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin[J]. EMBO J, 2004, 23(3): 489–499. DOI: 10.1038/sj.emboj.7600088 |
| [5] | MATTIROLI F, GU Y J, YADAV T, et al. DNA-mediated association of two histone-bound complexes of yeast Chromatin Assembly Factor-1(CAF-1) drives tetrasome assembly in the wake of DNA replication[J]. eLife, 2017, 6: e22799. DOI: 10.7554/eLife.22799 |
| [6] | ZHANG K, GAO Y, LI J J, et al. A DNA binding winged helix domain in CAF-1 functions with PCNA to stabilize CAF-1 at replication forks[J]. Nucleic Acids Res, 2016, 44(11): 5083–5094. DOI: 10.1093/nar/gkw106 |
| [7] | TANG Y, POUSTOVOITOV M V, ZHAO K H, et al. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly[J]. Nat Struct Mol Biol, 2006, 13(10): 921–929. DOI: 10.1038/nsmb1147 |
| [8] | TYLER J K, COLLINS K A, PRASAD-SINHA J, et al. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors[J]. Mol Cell Biol, 2001, 21(19): 6574–6584. DOI: 10.1128/MCB.21.19.6574-6584.2001 |
| [9] | ENGLISH C M, ADKINS M W, CARSON J J, et al. Structural basis for the histone chaperone activity of Asf1[J]. Cell, 2006, 127(3): 495–508. DOI: 10.1016/j.cell.2006.08.047 |
| [10] | TAGAMI H, RAY-GALLET D, ALMOUZNI G, et al. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis[J]. Cell, 2004, 116(1): 51–61. DOI: 10.1016/S0092-8674(03)01064-X |
| [11] | VERREAULT A, KAUFMAN P D, KOBAYASHI R, et al. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4[J]. Cell, 1996, 87(1): 95–104. DOI: 10.1016/S0092-8674(00)81326-4 |
| [12] | TAUNTON J, HASSIG C A, SCHREIBER S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p[J]. Science, 1996, 272(5260): 408–411. DOI: 10.1126/science.272.5260.408 |
| [13] | SONG J J, GARLICK J D, KINGSTON R E. Structural basis of histone H4 recognition by p55[J]. Genes Dev, 2008, 22(10): 1313–1318. DOI: 10.1101/gad.1653308 |
| [14] | SAUER P V, GU Y J, LIU W H, et al. Mechanistic insights into histone deposition and nucleosome assembly by the chromatin assembly factor-1[J]. Nucleic Acids Res, 2018, 46(19): 9907–9917. DOI: 10.1093/nar/gky823 |
| [15] | VOLK A, CRISPINO J D. The role of the chromatin assembly complex (CAF-1) and its p60 subunit (CHAF1b) in homeostasis and disease[J]. Biochim Biophys Acta Gene Regulat Mech, 2015, 1849(8): 979–986. DOI: 10.1016/j.bbagrm.2015.05.009 |
| [16] | LI Q, ZHOU H, WURTELE H, et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly[J]. Cell, 2008, 134(2): 244–255. DOI: 10.1016/j.cell.2008.06.018 |
| [17] | HUANG T H, FOWLER F, CHEN C C, et al. The histone chaperones ASF1 and CAF-1 promote MMS22L-TONSL-mediated Rad51 Loading onto ssDNA during homologous recombination in human cells[J]. Mol Cell, 2018, 69(5): 879–892. e5. DOI: 10.1016/j.molcel.2018.01.031 |
| [18] | TJEERTES J V, MILLER K M, JACKSON S P. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells[J]. EMBO J, 2009, 28(13): 1878–1889. DOI: 10.1038/emboj.2009.119 |
| [19] | VEMPATI R K, JAYANI R S, NOTANI D, et al. p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals[J]. J Biol Chem, 2010, 285(37): 28553–28564. DOI: 10.1074/jbc.M110.149393 |
| [20] | BURGESS R J, ZHOU H, HAN J H, et al. A role for Gcn5 in replication-coupled nucleosome assembly[J]. Mol Cell, 2010, 37(4): 469–480. DOI: 10.1016/j.molcel.2010.01.020 |
| [21] | ALLIS C D, CHICOINE L G, RICHMAN R, et al. Deposition-related histone acetylation in micronuclei of conjugating Tetrahymena[J]. Proc Natl Acad Sci U S A, 1985, 82(23): 8048–8052. DOI: 10.1073/pnas.82.23.8048 |
| [22] | SOBEL R E, COOK R G, ALLIS C D. Non-random acetylation of histone H4 by a cytoplasmic histone acetyltransferase as determined by novel methodology[J]. J Biol Chem, 1994, 269(28): 18576–18582. DOI: 10.1016/S0021-9258(17)32348-7 |
| [23] | SOBEL R E, COOK R G, PERRY C A, et al. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4[J]. Proc Natl Acad Sci U S A, 1995, 92(4): 1237–1241. DOI: 10.1073/pnas.92.4.1237 |
| [24] | PENG X D, FU H Y, YIN J J, et al. CHAF1B knockdown blocks migration in a hepatocellular carcinoma model[J]. Oncol Rep, 2018, 40(1): 405–413. |
| [25] | DUAN Y, LIU T Z, LI S W, et al. CHAF1B promotes proliferation and reduces apoptosis in 95-D lung cancer cells and predicts a poor prognosis in non-small cell lung cancer[J]. Oncol Rep, 2019, 41(4): 2518–2528. |
| [26] | YU Z S, LIU J Y, DENG W M, et al. Histone chaperone CAF-1:essential roles in multi-cellular organism development[J]. Cell Mol Life Sci, 2015, 72(2): 327–337. DOI: 10.1007/s00018-014-1748-3 |
| [27] | XU M, JIA Y L, LIU Z K, et al. Chromatin assembly factor 1, subunit A (P150) facilitates cell proliferation in human hepatocellular carcinoma[J]. Onco Targets Ther, 2016, 9: 4023–4035. DOI: 10.2147/OTT.S107050 |
| [28] | VOLK A, LIANG K W, SURANENI P, et al. A CHAF1B-dependent molecular switch in hema-topoiesis and leukemia pathogenesis[J]. Cancer cell, 2018, 34(5): 707–723. e7. DOI: 10.1016/j.ccell.2018.10.004 |
| [29] | DI M P, WANG M, MIAO J J, et al. CHAF1B induces radioresistance by promoting DNA damage repair in nasopharyngeal carcinoma[J]. Biomed Pharma-cother, 2020, 123: 109748. DOI: 10.1016/j.biopha.2019.109748 |
| [30] | BARBIERI E, DE PRETER K, CAPASSO M, et al. Histone chaperone CHAF1A inhibits differentiation and promotes aggressive neuroblastoma[J]. Cancer Res, 2014, 74(3): 765–774. DOI: 10.1158/0008-5472.CAN-13-1315 |
| [31] | CHELOUFI S, ELLING U, HOPFGARTNER B, et al. The histone chaperone CAF-1 safeguards somatic cell identity[J]. Nature, 2015, 528(7581): 218–224. DOI: 10.1038/nature15749 |
| [32] | YOUNG T J, CUI Y, PFEFFER C, et al. CAF-1 and Rtt101p function within the replication-coupled chromatin assembly network to promote H4 K16ac, preventing ectopic silencing[J]. PLoS Genet, 2020, 16(12): e1009226. DOI: 10.1371/journal.pgen.1009226 |
| [33] | HUANG H, YU Z S, ZHANG S Q, et al. Drosophila CAF-1 regulates HP1-mediated epigenetic silencing and pericentric heterochromatin stability[J]. J Cell Sci, 2010, 123(Pt 16): 2853–2861. |
| [34] | ROELENS B, CLÉMOT M, LEROUX-COYAU M, et al. Maintenance of heterochromatin by the large subunit of the CAF-1 replication-coupled histone chaperone requires its interaction with HP1a through a conserved motif[J]. Genetics, 2017, 205(1): 125–137. DOI: 10.1534/genetics.116.190785 |
| [35] | YANG B X, EL FARRAN C A, GUO H C, et al. Systematic identification of factors for provirus silencing in embryonic stem cells[J]. Cell, 2015, 163(1): 230–245. DOI: 10.1016/j.cell.2015.08.037 |
| [36] | NG C, AICHINGER M, NGUYEN T, et al. The histone chaperone CAF-1 cooperates with the DNA methyltransferases to maintain Cd4 silencing in cytotoxic T cells[J]. Genes Dev, 2019, 33(11-12): 669–683. DOI: 10.1101/gad.322024.118 |
| [37] | HATANAKA Y, INOUE K, OIKAWA M, et al. Histone chaperone CAF-1 mediates repressive histone modifications to protect preimplantation mouse embryos from endogenous retrotransposons[J]. Proc Natl Acad Sci U S A, 2015, 112(47): 14641–14646. DOI: 10.1073/pnas.1512775112 |
| [38] | JIANG D H, BERGER F. DNA replication-coupled histone modification maintains Polycomb gene silencing in plants[J]. Science, 2017, 357(6356): 1146–1149. DOI: 10.1126/science.aan4965 |
| [39] | CHENG L, ZHANG X, WANG Y, et al. Chromatin Assembly Factor 1(CAF-1) facilitates the establishment of facultative heterochromatin during pluripotency exit[J]. Nucleic Acids Res, 2019, 47(21): 11114–11131. DOI: 10.1093/nar/gkz858 |
| [40] | CHELOUFI S, HOCHEDLINGER K. Emerging roles of the histone chaperone CAF-1 in cellular plasticity[J]. Curr Opin Genet Dev, 2017, 46: 83–94. DOI: 10.1016/j.gde.2017.06.004 |
| [41] | ZHANG L, YU M Y, XU H Y, et al. RNA sequencing revealed the abnormal transcriptional profile in cloned bovine embryos[J]. Int J Biol Macromol, 2020, 150: 492–500. DOI: 10.1016/j.ijbiomac.2020.02.026 |
| [42] | AN Q L, PENG W, CHENG Y Y, et al. Melatonin supplementation during in vitro maturation of oocyte enhances subsequent development of bovine cloned embryos[J]. J Cell Physiol, 2019, 234(10): 17370–17381. DOI: 10.1002/jcp.28357 |
| [43] | ZHOU C, WANG Y Z, ZHANG J C, et al. H3K27 me3 is an epigenetic barrier while KDM6A overexpression improves nuclear reprogramming efficiency[J]. FASEB J, 2019, 33(3): 4638–4652. DOI: 10.1096/fj.201801887R |
| [44] |
梁素丽, 李向臣, 靳亚平, 等. 蛋白酶抑制剂MG-132对牛核移植重构胚重编程的影响[J]. 畜牧兽医学报, 2010, 41(12): 1536–1542.
LIANG S L, LI X C, JIN Y P, et al. Effects of protease inhibitor MG-132 on nuclear remodeling of bovine nuclear transfer reconstructed embryos[J]. Acta Veterinaria et Zootechnica Sinica, 2010, 41(12): 1536–1542. (in Chinese) |
| [45] | MATOBA S, LIU Y T, LU F L, et al. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation[J]. Cell, 2014, 159(4): 884–895. DOI: 10.1016/j.cell.2014.09.055 |
| [46] | ISHIUCHI T, ABE S, INOUE K, et al. Reprogramming of the histone H3.3 landscape in the early mouse embryo[J]. Nat Struct Mol Biol, 2021, 28(1): 38–49. DOI: 10.1038/s41594-020-00521-1 |
| [47] | ISHIUCHI T, ENRIQUEZ-GASCA R, MIZUTANI E, et al. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly[J]. Nat Struct Mol Biol, 2015, 22(9): 662–671. DOI: 10.1038/nsmb.3066 |
| [48] | CHUNG Y G, MATOBA S, LIU Y T, et al. Histone demethylase expression enhances human somatic cell nuclear transfer efficiency and promotes derivation of pluripotent stem cells[J]. Cell Stem Cell, 2015, 17(6): 758–766. DOI: 10.1016/j.stem.2015.10.001 |
| [49] | SAMPAIO R V, SANGALLI J R, DE BEM T H C, et al. Catalytic inhibition of H3K9 me2 writers disturbs epigenetic marks during bovine nuclear reprogram-ming[J]. Sci Rep, 2020, 10(1): 11493. DOI: 10.1038/s41598-020-67733-9 |
| [50] | PEASTON A E, EVSIKOV A V, GRABER J H, et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos[J]. Dev Cell, 2004, 7(4): 597–606. DOI: 10.1016/j.devcel.2004.09.004 |
| [51] | EVSIKOV A V, DE VRIES W N, PEASTON A E, et al. Systems biology of the 2-cell mouse embryo[J]. Cytogenet Genome Res, 2004, 105(2-4): 240–250. DOI: 10.1159/000078195 |
| [52] | PROBST A V, OKAMOTO I, CASANOVA M, et al. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development[J]. Dev Cell, 2010, 19(4): 625–638. DOI: 10.1016/j.devcel.2010.09.002 |
| [53] | GOMES A P, ILTER D, LOW V, et al. Dynamic incorporation of histone h3 variants into chromatin is essential for acquisition of aggressive traits and metastatic colonization[J]. Cancer cell, 2019, 36(4): 402–417. e13. DOI: 10.1016/j.ccell.2019.08.006 |
| [54] | WANG Y L, LI Y H, LUAN D J, et al. Dynamic replacement of H3.3 affects nuclear reprogramming in early bovine SCNT embryos[J]. Theriogenology, 2020, 154: 43–52. DOI: 10.1016/j.theriogenology.2020.05.031 |
| [55] | YAN H Y, XIANG X F, CHEN Q F, et al. HP1 cooperates with CAF-1 to compact heterochromatic transgene repeats in mammalian cells[J]. Sci Rep, 2018, 8(1): 14141. |
| [56] | FILIPESCU D, SZENKER E, ALMOUZNI G. Developmental roles of histone H3 variants and their chaperones[J]. Trends Genet, 2013, 29(11): 630–640. DOI: 10.1016/j.tig.2013.06.002 |
| [57] | HAKE S B, ALLIS C D. Histone H3 variants and their potential role in indexing mammalian genomes: the "H3 barcode hypothesis"[J]. Proc Natl Acad Sci U S A, 2006, 103(17): 6428–6435. DOI: 10.1073/pnas.0600803103 |
| [58] | LIU X, WANG Y Z, GAO Y P, et al. H3K9 demethylase KDM4E is an epigenetic regulator for bovine embryonic development and a defective factor for nuclear reprogramming[J]. Development, 2018, 145(4): dev158261. DOI: 10.1242/dev.158261 |
| [59] | AKIYAMA T, SUZUKI O, MATSUDA J, et al. Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos[J]. PLoS Genet, 2011, 7(10): e1002279. DOI: 10.1371/journal.pgen.1002279 |
| [60] | RAY-GALLET D, WOOLFE A, VASSIAS I, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity[J]. Mol Cell, 2011, 44(6): 928–941. |
| [61] | CHELOUFI S, ELLING U, HOPFGARTNER B, et al. The histone chaperone CAF-1 safeguards somatic cell identity[J]. Nature, 2015, 528(7581): 218–224. |
| [62] | SERRA-CARDONA A, ZHANG Z G. Replication-coupled nucleosome assembly in the passage of epigenetic information and cell identity[J]. Trends Biochem Sci, 2018, 43(2): 136–148. |



