2. 重庆市畜牧科学院, 重庆 402460;
3. 农业部养猪科学重点实验室, 重庆 402460
2. Chongqing Academy of Animal Science, Chongqing 402460, China;
3. Key Laboratory of Pig Industry Sciences, Ministry of Agriculture, Chongqing 402460, China
罗伊氏乳杆菌是一种具有很强特异性的益生菌,也是肠道微生物群的重要成员[1]。研究表明,罗伊氏乳杆菌对仔猪生长、肠道微生物菌群组成与代谢、肠道免疫功能等都具有重要的调节作用。早期补充罗伊氏乳杆菌有助于仔猪肠道菌群的稳定,减少肠道潜在病原体的数量,降低仔猪腹泻率[2-3];可以促进肠绒毛发育,提高肠道防御肽、紧密连接蛋白等的表达量,增加仔猪肠道屏障的完整性[4-6];还能通过Toll样受体信号通路抑制炎症因子的表达来减少肠道炎性反应[7],但具体的分子机制目前尚不十分清楚。
miRNA是一类参与真核和原核细胞转录调控的内源性非编码RNA分子,在肠上皮细胞分化和免疫调控中具有非常重要的作用[8-10]。它们不仅可以直接调控肠道免疫相关因子,而且可以通过影响肠道模式识别受体间接调控肠道免疫[11]。试验发现,外源微生物可以影响宿主肠道miRNA的表达,进而调控宿主肠道的健康。仔猪感染沙门菌后,回肠miR-29a显著上调,导致小窝蛋白2显著下调[12];嗜酸乳杆菌和双歧杆菌能降低小鼠结肠中癌基因miR-135b、miR-155的表达,增加肿瘤抑制基因miR-26b、miR-18a的表达,从而改善结肠癌[13]。然而,对于肠道miRNA是否参与罗伊氏乳杆菌对仔猪肠道健康的调节过程,目前尚未见相关报道。因此,本试验拟通过高通量测序,研究罗伊氏乳杆菌对仔猪空肠miRNA表达谱的影响,探寻发生明显变化的肠道miRNAs并预测可能的作用通路,为深入探究罗伊氏乳杆菌在肠道上的作用机制提供试验依据。
1 材料与方法 1.1 试验材料和样品采集选用1日龄健康的约克×荣昌仔猪6窝,随机分为对照组和试验组,每组3窝(共27头)。从出生开始,对照组每天每头灌喂2 mL 0.1%的蛋白胨溶液,LR组每天每头灌喂2 mL 0.1%蛋白胨溶液溶解的罗伊氏乳杆菌(罗伊氏乳杆菌I5007 0.5×1010 CFU·mL-1,由中国农业大学提供),连续灌喂20 d。试验期间,哺乳母猪自由采食和饮水,仔猪由各自母猪哺乳喂养,自由饮水;母、仔猪按常规程序进行管理。在第10日龄(对照组CN10、LR组LR10)和20日龄(对照组CN20、LR组LR20)时,每组选取接近平均体重的6头仔猪(2头·窝-1)进行屠宰,采集空肠中段样品,于液氮迅速冻存后再转移至-80 ℃冰箱中保存备用。
1.2 RNA文库构建及高通量测序空肠组织液氮研磨,用Trizol试剂提取总RNA,每组6个重复,3个重复等量混合为一个样,共2个样,检测混合后的总RNA质量,保证样品RIN为7.5~9.0。测序流程:15%的聚丙烯酰胺凝胶电泳分离长度为18~30 nt的小RNA,连接3′和5′接头,然后进行反转录,合成cDNA,再进行PCR扩增,最后凝胶电泳并纯化,由此构建对照组CN10、CN20和LR组LR10、LR20文库,每个组2个重复,共建8个文库,分别为CN10a、CN10b、CN20a、CN20b、LR10a、LR10b、LR20a和LR20b。文库构建完成后,上机测序,使用BGISEQ-500测序平台进行高通量测序,测序由华大基因科技有限公司完成。
1.3 鉴定miRNA序列及miRNA的差异表达分析测序所得序列去除低质量、接头污染和长度小于18 nt等序列,然后统计小RNA片段的序列总数、种类和长度分布,获得clean reads。通过SOAP和bowtie筛选后的sRNA与猪的参考基因定位,去除rRNA、tRNA、scRNA、snRNA、snoRNA等小RNA,过滤后,与miRBase21.0中猪的已知miRNAs进行匹配,匹配上的序列为已知miRNAs,再用mirdeep软件预测新miRNA。对样本中已知miRNA的表达量进行统计,用TPM进行表达量归一化处理。使用SPSS 18.0中的描述统计分析数据,再使用ExpDiff方法比较对照组和试验组之间miRNA表达量的差异,计算对照组和试验组中miRNA的差异倍数(Fold change)和P值,筛选出差异表达的miRNAs。
1.4 靶基因预测和功能分析用RNAhybrid、miRanda和TargetScan多个软件对差异表达的已知miRNAs进行靶基因预测,取交集作为预测结果。获得靶基因后,与KEGG通路公共数据库进行匹配,找出与整个基因组背景相比显著富集的通路(P < 0.05),并确定显著富集通路的靶基因数目。KEGG通路的分析有助于更进一步了解基因的生物学功能,确定候选基因参与的最主要生化代谢途径和信号转导途径。
1.5 差异极显著miRNAs验证为验证高通量测序结果的准确性,随机挑选10和20日龄差异表达的miRNAs各3个,每个样品设置3个重复,进行荧光定量PCR(qRT-PCR)试验。首先,根据选中miRNA的成熟序列设计荧光定量PCR引物(表 1),然后,利用反转录试剂盒SYBR Prime Script RM miRNA RT-PCR获得cDNA,以U6作为内参基因,用SYBR Premix Ex TaqTM II试剂盒进行荧光定量PCR反应,反应体系:cDNA模板2.0 μL、miRNA引物0.8 μL、下游引物(试剂盒)0.8 μL、2×SYBR Premix Ex Taq II 10 μL、ROX Reference Dye 0.4 μL、ddH2O 7 μL;反应条件为:1)95 ℃变性10 s;2)扩增:95 ℃,5 s;60 ℃,20 s;40个循环。所有的反应设置3个重复,结果采用2-△△CT法进行相对定量统计分析。
|
|
表 1 荧光定量PCR引物序列 Table 1 The primer sequences of qRT-PCR |
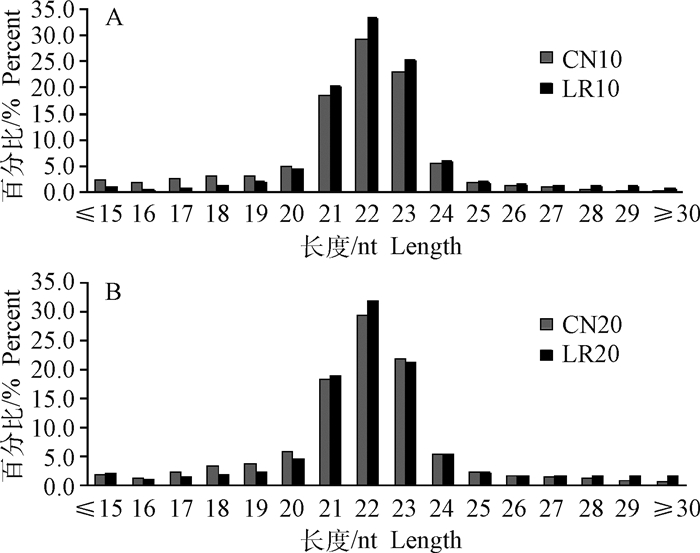
经过Solexa测序得到raw data数据,进行过滤处理,主要包括去除接头、污染及低质量等序列。本试验共建了8个文库,每个试验组包含a和b两个文库(例如CN10组包括CN10a和CN10b),为保证后续分析结果的准确性,每个文库截取完全一样的raw reads进行过滤分析。CN10共获得了47 207 231条clean reads,占两个文库raw reads总数的93.80%;CN20获得了47 156 868条clean reads,占两个文库raw reads总数的93.69%;LR10获得了47 744 290条clean reads,占两个文库raw reads总数的94.86%;LR20获得了44 995 451条clean reads,占两个文库raw reads总数的89.40%(表 2)。筛选对照组和LR组获得的纯净序列,选取一定长度范围内的小RNA进行长度统计分布,过滤掉小于18 nt、大于30 nt的reads,绝大多数序列长度为20~24 nt,其中22 nt长度序列占的比例最大(图 1)。
|
|
表 2 数据过滤信息 Table 2 Data filtering information |
|
图 1 10(A)和20日龄(B)小RNA序列长度分布图 Fig. 1 Length distribution of small RNA sequences at 10(A) and 20(B) days of age |
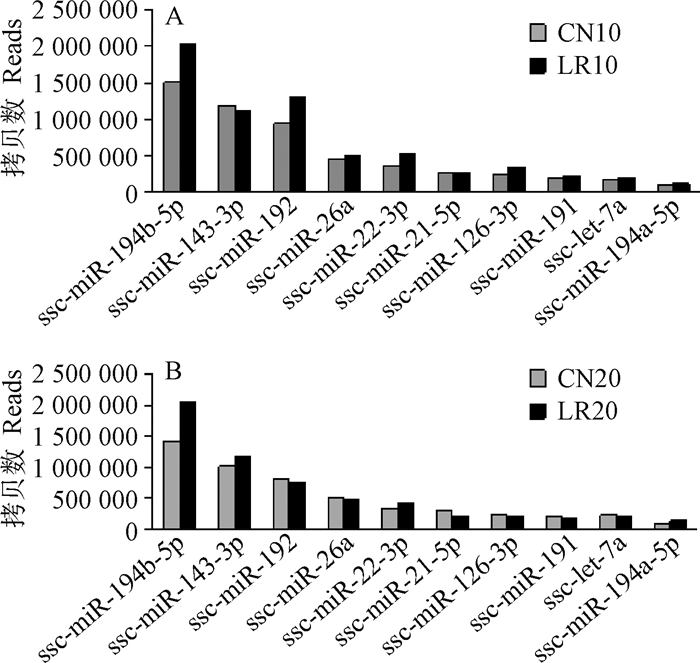
通过与miRBase 21.0中猪miRNAs序列比对,10日龄鉴定出354个已知的miRNAs,20日龄鉴定出352个已知的miRNAs,有329个miRNAs在两个日龄共表达。其中miR-194b-5p、miR-192、miR-143-3p、miR-26a、miR-22-3p和miR-21-5p等miRNAs在两个日龄的空肠组织中都高表达,表达量都在100 000拷贝数以上,与肠道发育密切相关的miR-194b-5p和miR-26a表达量均排在前五(图 2)。
|
图 2 10(A)和20日龄(B)表达量前10的miRNAs Fig. 2 The top 10 expressed miRNAs at 10(A) and 20(B) days of age |
将对照组和LR组的已知miRNAs用软件进行统计分析,以log2(Fold change)>1或 < -1(正值代表上调,负值代表下调),P < 0.05为标准筛选两个组之间差异表达的miRNAs(表 3),分析得出,20日龄差异表达的miRNAs数量比10日龄的多2个。在10日龄有7个差异表达的miRNAs,3个上调,4个下调,其中ssc-miR-218b差异倍数最大;而ssc-miR-150虽然表达量最高,但差异倍数较小。在20日龄有9个差异表达的miRNAs,5个上调,4个下调,ssc-miR-182和ssc-miR-125a表达量最高,但差异倍数都不大。在差异表达的miRNAs中,只有ssc-miR-218在两个日龄中都差异表达(表 3)。
|
|
表 3 灌喂罗伊氏乳杆菌后空肠组织显著差异表达的miRNAs Table 3 Significant differentially expressed miRNAs in the jejunum response to L. reuteri treatment |
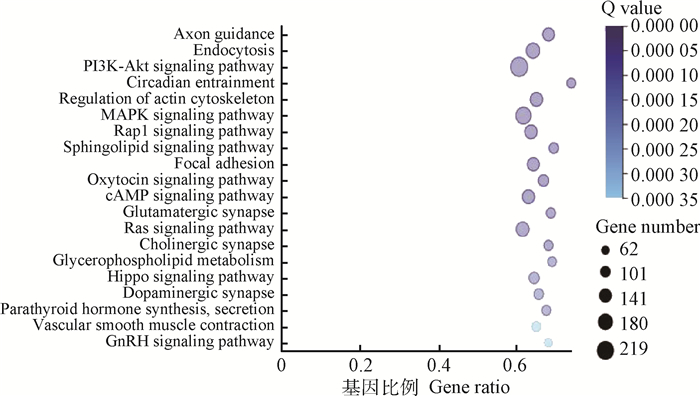
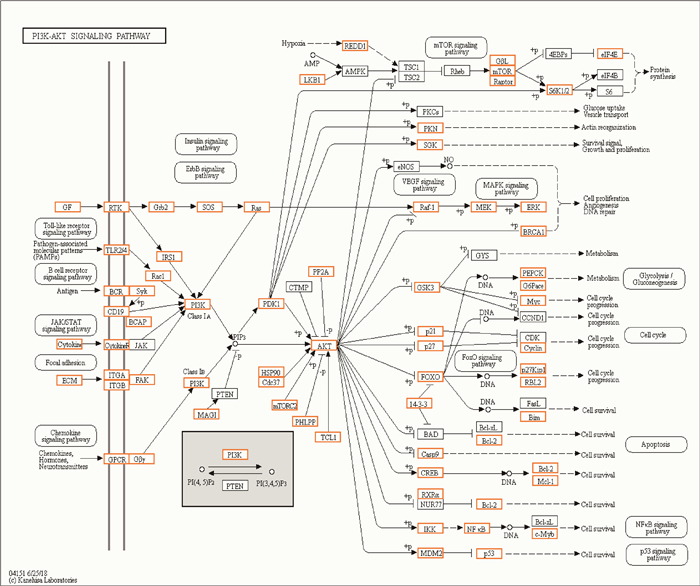
10和20日龄的空肠分别有7和9个miRNAs差异表达显著,只有1个miRNA在两个日龄中都出现差异表达,故共有15个差异表达的miRNAs。用RNAhybrid、miRanda和TargetScan分析软件对其进行靶基因预测,共得到10 221个靶基因。KEGG功能分析发现,靶基因共富集到293条通路中,其中有66条显著富集通路(P < 0.05),图 3中列出前20个显著富集通路。在显著富集的通路中,前5个靶基因数量最多的通路是PI3K-Akt信号通路(219个靶基因)、MAPK信号通路(184个靶基因)、胞吞作用(156个靶基因)、Ras信号通路(144个靶基因)和cAMP信号通路(133个靶基因),这些通路调节着细胞的凋亡、生长以及一些重要基因的表达,根据信号通路中包含的靶基因及可能的生物学意义,其中被证实参与调节宿主肠道发育及免疫调节的PI3K-Akt和MAPK信号通路,可能参与调节仔猪肠道健康。在PI3K-Akt信号通路中(图 4),Toll样受体和MAPK等免疫相关信号通路参与其中;而在MAPK信号通路中(图 5),与炎症免疫反应相关的TNF信号通路也参与其中。在两个信号通路中,抑癌基因p53,Ras和涉及机体防御反应相关的因子NF-κB等都是重要的靶基因。
|
图 3 15个差异表达miRNAs靶基因富集的前20位KEGG通路 Fig. 3 The top 20 KEGG pathways enriched for target genes of the 15 differentially expressed miRNAs |
|
图 4 PI3K-Akt信号通路 Fig. 4 PI3K-Akt signaling pathway |

|
图 5 MAPK信号通路 Fig. 5 MAPK signaling pathway |
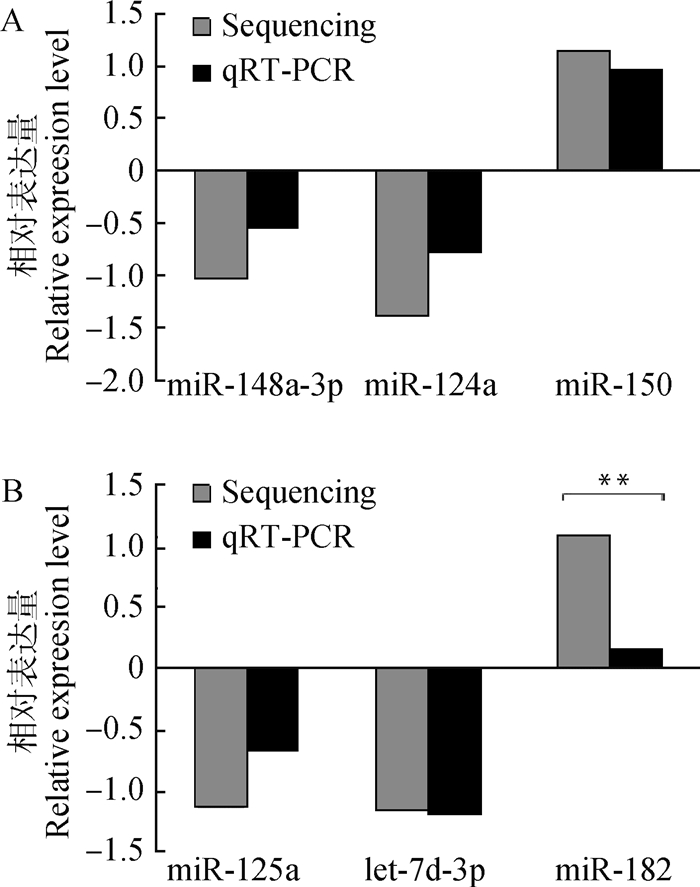
在15个差异表达的miRNAs中,10和20日龄各随机挑选3个miRNAs进行qRT-PCR验证,在10日龄中检测ssc-miR-124a、ssc-miR-150和ssc-miR-148a-3p的表达,在20日龄中检测ssc-miR-125a、ssc-miR-182和ssc-let-7d-3p的表达。qRT-PCR结果以2-△△CT表示,与测序结果的log2 (Fold change)值相对比。图 6中,qRT-PCR的测定结果与Solexa测序结果基本一致,6个miRNAs在空肠组织中都有表达,仅在第20日龄中,ssc-miRNA-182的表达与测序结果相比显著降低(P < 0.01)。
|
**表示差异极显著(P<0.01) ** indicates highly significant difference (P < 0.01) 图 6 10(A)和20日龄(B)中随机挑选6个差异表达miRNAs的qRT-PCR验证 Fig. 6 qRT-PCR verification of the differentially expressed miRNAs randomly seleted from 10-day-old (A) and 20-day-old (B) |
miRNAs已经被证实是调节宿主肠道基因表达的一个重要因素,且在肠道微生物与宿主互作中,参与宿主细胞的分化、免疫应答等过程[14]。本文统计了仔猪10和20日龄空肠中表达量前10的miRNAs,miR-194b-5p、miR-21-5p、miR-143-3p和miR-26a在仔猪空肠中高表达,这与Sharbati等[15]在仔猪小肠上的研究结果一致,其中miR-194b-5p能促进肠上皮细胞的分化[16];而miR-22-3p、miR-143-3p、miR-192和let-7a被证实与胃肠道癌有关[17-18]。以上结果提示,这些高表达的miRNAs可能在仔猪肠道中发挥着重要功能。
本试验比较分析了有/无灌喂罗伊氏乳杆菌对仔猪空肠miRNA表达谱的影响,发现灌喂罗伊氏乳杆菌后,仔猪空肠有15个miRNAs差异表达显著(P < 0.05),在差异表达的miRNAs中,miR-221-3p、miR-150、miR-130b-5p、miR-9、miR-124a、miR-218和miR-182被证实参与胃肠道癌的调节[19-22],而miR-423-5p、miR-125a和miR-490-5p则被发现与肠道炎症相关[23-24]。试验已证实,miR-423-5p、miR-218和miR-150对肠道的健康具有重要的调控作用,miR-423-5p是三叶因子1的靶标,参与调节免疫球蛋白和细胞之间的相互作用[25]。本试验发现,给新生仔猪灌喂罗伊氏乳杆菌使空肠miR-423-5p显著上调,这与Kreuzer-Redmer等[26]的试验结果类似,他们给仔猪饲喂屎肠球菌后,回肠miR-423-5p也显著上调,并通过抑制免疫球蛋白λ轻链C区的合成,影响仔猪肠道免疫功能。miR-218主要调节细胞增殖、分化和凋亡,其过表达能抑制结肠癌和直肠癌的发生[27];miR-150通过调节体内β-连环蛋白抑制结直肠癌的生长[28]。在本试验中,仔猪空肠miR-218和miR-150显著上调。推测罗伊氏乳杆菌可能通过影响上述差异表达的miRNAs来调控仔猪肠道健康。
生物信息学分析发现,15个差异表达miRNAs的靶基因显著富集到66条通路中(P < 0.05),其中,PI3K-Akt和MAPK信号通路在益生菌调节宿主肠道中起重要作用。PI3K-Akt和MAPK信号通路涉及多种细胞功能,包括细胞增殖、分化和迁移,且能被多种类型的刺激所激活,MAPK信号通路还能响应促炎性刺激而激活细胞外调节蛋白激酶ERK1/2[29-33]。研究发现,罗伊氏乳杆菌能抑制脂多糖诱导的小鼠肠道MAPK和ERK1/2磷酸化信号的激活,下调促炎因子TNF-α、IL-6等的表达水平,进而调节肠道免疫反应和代谢,发挥益生作用[34];体外试验发现,罗伊氏乳杆菌在脂多糖处理的小鼠巨噬细胞中能够调节NF-κB的表达,并抑制PI3K-Akt途径,显著降低促炎因子TNF-α、IL-1β和IL-6的表达[35]。研究表明,miR-423-5p通过PI3K-Akt和MAPK信号转导途径来保护糖尿病、肾病中高糖介导的足细胞损伤[36];miR-218通过PI3K-Akt信号通路直接靶向肿瘤基因调节因子BMI-1,抑制癌细胞的增殖、迁移和侵袭能力[37];miR-124a通过抑制PI3K-Akt信号通路关键因子的表达,调节成纤维细胞样滑膜细胞增殖、分化,并抑制促炎因子TNF-α和IL-6的表达[38]。本课题组研究还发现,新生仔猪灌喂罗伊氏乳杆菌后,空肠促炎因子TNF-α、IL-1β和IL-6表达也都下调(数据未发表)。结合本试验结果提示,罗伊氏乳杆菌可能通过这些差异表达miRNAs介导的MAPK和PI3K-Akt信号通路来影响仔猪肠道的健康,但具体分子机制仍需后续验证。
4 结论通过高通量测序及其生物信息学分析获得罗伊氏乳杆菌作用仔猪空肠后的miRNA表达谱,筛选出10和20日龄仔猪空肠组织显著差异表达的15个miRNAs,这些miRNAs的靶基因预测和功能分析得出,MAPK和PI3K-Akt等信号通路可能参与罗伊氏乳杆菌调控的仔猪肠道健康。
| [1] | WEGMANN U, MACKENZIE D A, ZHENG J S, et al. The pan-genome of Lactobacillus reuteri strains originating from the pig gastrointestinal tract[J]. BMC Genomics, 2015, 16(1): 1023. DOI: 10.1186/s12864-015-2216-7 |
| [2] | LIU H, ZHANG J, ZHANG S H, et al. Oral administration of Lactobacillus fermentum I5007 favors intestinal development and alters the intestinal microbiota in formula-fed piglets[J]. J Agric Food Chem, 2014, 62(4): 860–866. |
| [3] | DOWARAH R, VERMA A K, AGARWAL N, et al. Effect of swine based probiotic on performance, diarrhoea scores, intestinal microbiota and gut health of grower-finisher crossbred pigs[J]. Livest Sci, 2017, 195: 74–79. DOI: 10.1016/j.livsci.2016.11.006 |
| [4] | TALARICO T L, CASAS I A, CHUNG T C, et al. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri[J]. Antimicrob Agents Chemother, 1988, 32(12): 1854–1858. DOI: 10.1128/AAC.32.12.1854 |
| [5] | HOU C L, WANG Q W, ZENG X F, et al. Complete genome sequence of Lactobacillus reuteri I5007, a probiotic strain isolated from healthy piglet[J]. J Biotechnol, 2014, 179: 63–64. DOI: 10.1016/j.jbiotec.2014.03.019 |
| [6] | YANG F J, WANG A N, ZENG X F, et al. Lactobacillus reuteri I5007 modulates tight junction protein expression in IPEC-J2 cells with LPS stimulation and in newborn piglets under normal conditions[J]. BMC Microbiol, 2015, 15(1): 32. |
| [7] | LIU Y Y, FATHEREE N Y, MANGALAT N, et al. Lactobacillus reuteri strains reduce incidence and severity of experimental necrotizing enterocolitis via modulation of TLR4 and NF-κB signaling in the intestine[J]. Am J Physiol Gastrointest Liver Physiol, 2012, 302(6): G608–G617. DOI: 10.1152/ajpgi.00266.2011 |
| [8] | PUA H H, ANSEL K M. MicroRNA regulation of allergic inflammation and asthma[J]. Curr Opin Immunol, 2015, 36: 101–108. DOI: 10.1016/j.coi.2015.07.006 |
| [9] | BARTEL D P. MicroRNAs:target recognition and regulatory functions[J]. Cell, 2009, 136(2): 215–233. DOI: 10.1016/j.cell.2009.01.002 |
| [10] | WU L Y, MA X P, SHI Y, et al. Alterations in microRNA expression profiles in inflamed and non-inflamed ascending colon mucosae of patients with active Crohn's disease[J]. J Gastroenterol Hepatol, 2017, 32(10): 1706–1715. DOI: 10.1111/jgh.13778 |
| [11] | MCKENNA L B, SCHUG J, VOUREKAS A, et al. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function[J]. Gastroenterology, 2010, 139(5): 1654–1664. DOI: 10.1053/j.gastro.2010.07.040 |
| [12] | HOEKE L, SHARBATI J, PAWAR K, et al. Intestinal Salmonella typhimurium infection leads to miR-29a induced Caveolin 2 regulation[J]. PLoS One, 2013, 8(6): e67300. DOI: 10.1371/journal.pone.0067300 |
| [13] | HEYDARI Z, RAHAIE M, ALIZADEH A M, et al. Effects of Lactobacillus acidophilus and Bifidobacterium bifidum probiotics on the expression of MicroRNAs 135b, 26b, 18a and 155, and their involving genes in mice colon cancer[J]. Probiotics Antimicrob Proteins, 2019, 11(4): 1155–1162. DOI: 10.1007/s12602-018-9478-8 |
| [14] |
孙倩, 王琪, 王敬, 等. miRNAs参与肠道微生物和宿主免疫互作的调控[J]. 动物营养学报, 2020, 32(1): 79–84.
SUN Q, WANG Q, WANG J, et al. miRNAs regulation on interaction between gut microbiota and host immunity[J]. Chinese Journal of Animal Nutrition, 2020, 32(1): 79–84. (in Chinese) |
| [15] | SHARBATI S, FRIEDLANDER M R, SHARBATI J, et al. Deciphering the porcine intestinal microRNA transcriptome[J]. BMC Genomics, 2010, 11(1): 275. DOI: 10.1186/1471-2164-11-275 |
| [16] | TAO X, XU Z W, MEN X M. Analysis of serum microRNA expression profiles and comparison with small intestinal microRNA expression profiles in weaned piglets[J]. PLoS One, 2016, 11(9): e0162776. DOI: 10.1371/journal.pone.0162776 |
| [17] | ARDILA H J, SANABRIA-SALAS M C, MENESES X, et al. Circulating miR-141-3p, miR-143-3p and miR-200c-3p are differentially expressed in colorectal cancer and advanced adenomas[J]. Mol Clin Oncol, 2019, 11(2): 201–207. |
| [18] | KONNO M, KOSEKI J, ASAI A, et al. Distinct methy-lation levels of mature microRNAs in gastrointestinal cancers[J]. Nat Commun, 2019, 10(1): 3888. |
| [19] | ZHANG Y, HUANG H F, ZHANG Y, et al. Combined detection of serum MiR-221-3p and MiR-122-5p expression in diagnosis and prognosis of gastric cancer[J]. J Gastric Cancer, 2019, 19(3): 315–328. DOI: 10.5230/jgc.2019.19.e28 |
| [20] | CHEN H, YANG Y Q, WANG J, et al. MiR-130b-5p promotes proliferation, migration and invasion of gastric cancer cells via targeting RASAL1[J]. Oncol Lett, 2018, 15(5): 6361–6367. |
| [21] | HANG C, YAN H S, GONG C, et al. MicroRNA-9 inhibits gastric cancer cell proliferation and migration by targeting neuropilin-1[J]. Exp Ther Med, 2019, 18(4): 2524–2530. |
| [22] | TAHARA T, TAHARA S, HORIGUCHI N, et al. Gastric mucosal microarchitectures associated with irreversibility with Helicobacter pylori eradication and downregulation of Micro RNA (miR)-124a[J]. Cancer Invest, 2019, 37(9): 417–426. DOI: 10.1080/07357907.2019.1663207 |
| [23] | SUN C M, WU J, ZHANG H, et al. Circulating miR-125a but not miR-125b is decreased in active disease status and negatively correlates with disease severity as well as inflammatory cytokines in patients with Crohn's disease[J]. World J Gastroenterol, 2017, 23(44): 7888–7898. DOI: 10.3748/wjg.v23.i44.7888 |
| [24] | REN H X, ZHANG F C, LUO H S, et al. Role of mast cell-miR-490-5p in irritable bowel syndrome[J]. World J Gastroenterol, 2017, 23(1): 93–102. DOI: 10.3748/wjg.v23.i1.93 |
| [25] | YOUNGER S T, COREY D R. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters[J]. Nucleic Acids Res, 2011, 39(13): 5682–5691. DOI: 10.1093/nar/gkr155 |
| [26] | KREUZER-REDMER S, BEKURTZ J C, ARENDS D, et al. Feeding of Enterococcus faecium NCIMB 10415 leads to intestinal miRNA-423-5p-induced regulation of immune-relevant genes[J]. Appl Environ Microbiol, 2016, 82(8): 2263–2269. DOI: 10.1128/AEM.04044-15 |
| [27] | LAMPROPOULOU D I, ARAVANTINOS G, LASCHOS K, et al. MiR-218 and miR-100 polymorphisms as markers of irinotecan-based chemotherapy response in metastatic colorectal cancer[J]. Int J Colorectal Dis, 2019, 34(11): 1871–1877. DOI: 10.1007/s00384-019-03401-3 |
| [28] | HE Z Y, DANG J, SONG A L, et al. The involvement of miR-150/β-catenin axis in colorectal cancer progression[J]. Biomed Pharmacother, 2020, 121: 109495. DOI: 10.1016/j.biopha.2019.109495 |
| [29] | KWON K H, OHIGASHI H, MURAKAMI A. Dextran sulfate sodium enhances interleukin-1β release via activation of p38 MAPK and ERK1/2 pathways in murine peritoneal macrophages[J]. Life Sci, 2007, 81(5): 362–371. DOI: 10.1016/j.lfs.2007.05.022 |
| [30] | WAETZIG G H, SEEGERT D, ROSENSTIEL P, et al. P38 mitogen-activated protein kinase is activated and linked to TNF-α signaling in inflammatory bowel disease[J]. J.Immunol, 2002, 168(10): 5342–5351. DOI: 10.4049/jimmunol.168.10.5342 |
| [31] | NAGAE K, UCHI H, MORINO-KOGA S, et al. Glucagon-like peptide-1 analogue liraglutide facilitates wound healing by activating PI3K/Akt pathway in keratinocytes[J]. Diabetes Res Clin Pract, 2018, 146: 155–161. DOI: 10.1016/j.diabres.2018.10.013 |
| [32] | LING L, GU S H, CHENG Y, et al. bFGF promotes Sca-1+ cardiac stem cell migration through activation of the PI3K/Akt pathway[J]. Mol Med Rep, 2018, 17(2): 2349–2356. |
| [33] | LV C, YANG S W, CHEN X, et al. MicroRNA-21 promotes bone mesenchymal stem cells migration in vitro by activating PI3K/Akt/MMPs pathway[J]. J Clin Neurosci, 2017, 46: 156–162. DOI: 10.1016/j.jocn.2017.07.040 |
| [34] | GAO K, LIU L, DOU X X, et al. Doses Lactobacillus reuteri depend on adhesive ability to modulate the intestinal immune response and metabolism in mice challenged with lipopolysaccharide[J]. Sci Rep, 2016, 6: 28332. DOI: 10.1038/srep28332 |
| [35] | GRIET M, ZELAYA H, MATEOS M V, et al. Soluble factors from Lactobacillus reuteri CRL1098 have anti-inflammatory effects in acute lung injury induced by lipopolysaccharide in mice[J]. PLoS One, 2014, 9(10): e110027. DOI: 10.1371/journal.pone.0110027 |
| [36] | XU Y X, ZHANG J Z, FAN L, et al. miR-423-5p suppresses high-glucose-induced podocyte injury by targeting Nox4[J]. Biochem Biophys Res Commun, 2018, 505(2): 339–345. DOI: 10.1016/j.bbrc.2018.09.067 |
| [37] | XU L, SUN H B, XU Z N, et al. MicroRNA-218 regulates the epithelial-to-mesenchymal transition and the PI3K/Akt signaling pathway to suppress lung adenocarcinoma progression by directly targeting BMI-1[J]. Eur Rev Med Pharmacol Sci, 2019, 23(18): 7978–7988. |
| [38] | YANG B L, GE Y, ZHOU Y, et al. MiR-124a inhibits the proliferation and inflammation in rheumatoid arthritis fibroblast-like synoviocytes via targeting PIK3/NF-κB pathway[J]. Cell Biochem Funct, 2019, 37(4): 208–215. DOI: 10.1002/cbf.3386 |



