2. 河南农业大学 禽病研究所, 郑州 450046
2. Institute of Poultry Diseases, Henan Agricultural University, Zhengzhou 450046, China
鸭源鸡杆菌(Gallibacterium anatis, G. anatis)属于巴氏杆菌科鸡杆菌属,包括溶血型和非溶血型[1-2]。溶血型鸭源鸡杆菌在禽类中普遍流行,其感染往往造成蛋鸡输卵管炎和腹膜炎,产蛋率下降和死亡率升高[3-5]。而且,国内流行菌株普遍存在多重耐药[6-7],药物防制效果差。随着我国限制抗生素在畜牧业中的使用,创制安全有效的疫苗是疾病防控研究的首选,而全面了解其发病机制是当务之急。尽管菌毛[8]、金属蛋白酶[9]、RTX毒素GtxA[10-11]、血凝素[12-13]、外膜囊泡[14]等多种毒力因子得到鉴定,鸭源鸡杆菌外膜蛋白(outer membrane proteins,OMPs)的功能还罕见研究。王艳等[15]对外膜蛋白A(outer membrane protein A, OmpA)进行原核表达,并作了免疫保护性分析。而外膜蛋白W(outer membrane protein W, OmpW)目前仅笔者报道其与鸭源鸡杆菌耐药性相关[16],外膜蛋白其他生物学功能还有待研究。
OmpW是细菌外膜蛋白家族成员之一,属于小外膜蛋白家族,在革兰阴性菌中普遍存在[17]。该蛋白是多种细菌应对渗透应激的一种重要外膜蛋白,副溶血性弧菌[18]、嗜水气单胞菌[19]、美人鱼发光杆菌[20]、坂崎肠杆菌[21-22]和霍乱弧菌[23]的外膜蛋白在抗渗透应激中发挥重要作用。探究OmpW在鸭源鸡杆菌抗渗透应激中扮演的角色对于了解该菌致病机制具有一定意义。本研究首次分析了鸭源鸡杆菌及其ompW缺失株ΔompW在渗透应激环境中的生存能力和生长性能,并从转录水平和蛋白水平分析了两者在不同盐度下外膜蛋白的表达差异,以期了解OmpW在鸭源鸡杆菌抗渗透应激中的作用。
1 材料与方法 1.1 菌株鸭源鸡杆菌PDS-RZ-1-SLG株(RZ)分离自河南某鸡场患输卵管炎的蛋鸡,由本实验室分离并保存[24];鸭源鸡杆菌OmpW缺失株ΔompW由本课题组构建、保存[16]。
1.2 主要试剂BHI培养基购自Difco公司;无维生素酪蛋白氨基酸购自美国BD公司;cDNA第一链合成试剂盒、SYBR Green Real-time PCR Premix Ex Taq Ⅱ、预染蛋白质Marker、低分子量蛋白质Marker购自宝生物工程(大连)有限公司(TaKaRa);Na-K ATPase多抗购自Sigma公司。
1.3 引物基于鸭源鸡杆菌RZ株基因组设计ompW基因和16S rRNA的实时定量PCR引物,引物序列分别如下:TTAGCACTAGCACTTGCAACTTC(ompW-F)/ TGCATTACTGTTTACATCAAAATCA (ompW-R)和AGAACCTTACCTACTCTTGAC(16S rRNA-F)/TAACCCAACATTTCACAACAC(16S rRNA-R),预期扩增片段的大小分别为160和122 bp。引物由生工生物工程(上海)股份有限公司合成。
1.4 不同盐度中鸭源鸡杆菌生长性能测定按照文献[25]描述的方法分析鸭源鸡杆菌RZ和ΔompW在渗透应激(0.5%、3.0% NaCl)中的生存差异。用平板活菌计数法确定菌液浓度。菌株应激耐受率用应激处理组菌落浓度(CFU·mL-1)/对照组菌液浓度(CFU·mL-1)×100%表示。每个样品3个重复,试验重复3遍。
参照文献[16]描述的方法测定鸭源鸡杆菌RZ和ΔompW在正常(0.5% NaCl)和高盐(3.0% NaCl)BHI液体培养基中的生长性能。试验设三次重复,取平均值。运用软件GraphPad Prism 6绘制细菌生长曲线。
1.5 不同盐度中鸭源鸡杆菌OMPs的表达分析挑取RZ和ΔompW单菌落分别接种于含5%血清的BHI液体培养基中,37 ℃过夜振荡培养。第2天,无菌PBS洗涤2遍后将菌液分别调至OD600 mm=0.5。按1:100分别接种于含0.5%、2.0%、3.0% NaCl的BHI液体培养基中,37 ℃振荡培养2 h。按照Kim等[26]描述的超速离心法提取外膜蛋白,取每份样品20 μg进行SDS-PAGE。同时用Western blot分析鸭源鸡杆菌在正常(0.5% NaCl)和高盐(3.0% NaCl)BHI液体培养基中OmpW的表达差异(自制兔抗OmpW多克隆抗体作为一抗)。用Na-K ATPase作为内参。
1.6 实时定量PCR检测不同盐度下OmpW mRNA表达水平用细菌RNA提取试剂盒提取“1.5”中0.5%、3.0% NaCl BHI中培养的为鸭源鸡杆菌(OD600 nm=0.5)总RNA。按照cDNA第一链合成试剂盒说明书合成cDNA,使用SYBR Premix Ex Taq Ⅱ试剂盒进行实时定量PCR,检测OmpW mRNA的表达水平。反应体系为20 μL:2×SYBR Premix Ex Taq Ⅱ 10 μL,上、下游引物(10 μmol·L-1)各0.5 μL,cDNA 2 μL(< 100 ng),ddH2O 7 μL。反应条件:95 ℃预变性2 min,95 ℃变性20 s、56 ℃退火30 s,40个循环,得到各样品的Ct值。利用16S rRNA基因作为内参基因,用2-△△Ct法分析OmpW mRNA相对表达水平。试验进行三次重复。
1.7 数据分析试验数据采用“
用生存率表示细菌的耐受渗透应激的能力。在高盐(3.0% NaCl)渗透应激下,ΔompW明显低于RZ的生存率(如图 1,83.50% vs 95.17%), 且差异显著(P < 0.05)。

|
过夜培养的菌液,无菌PBS洗涤2遍后用PBS调整其OD600 nm=1.5。在含3% NaCl BHI中孵育1 h,同时设置阴性对照。细菌耐应激生存率用处理样品细菌浓度/对照组细菌浓度×100%表示。*.P < 0.05 Overnight cultures were washed twice in sterile PBS and diluted to an OD600 nm value of 1.5. Then cell suspensions were cultured in BHI of 3% NaCl for 1 h. Cell suspension without stress as negative control. The survival rate of stress-resistant cells was calculated as (stressed sample CFU·mL-1)/(control sample CFU·mL-1)×100%.*.P < 0.05 图 1 鸭源鸡杆菌RZ和ΔompW渗透应激抵抗能力分析 Fig. 1 Analysis of the osmotic stress resistance of G. anatis RZ and ΔompW |
与正常盐度培养基中的生长情况相比,RZ和ΔompW在高盐度培养基(3.0% NaCl)中生长速度均下降,且对数生长期起始时间被推迟。而且,在高盐度培养基(3.0% NaCl)中RZ生长速度始终高于ΔompW的生长速度,差异不显著(P>0.5,图 2)。
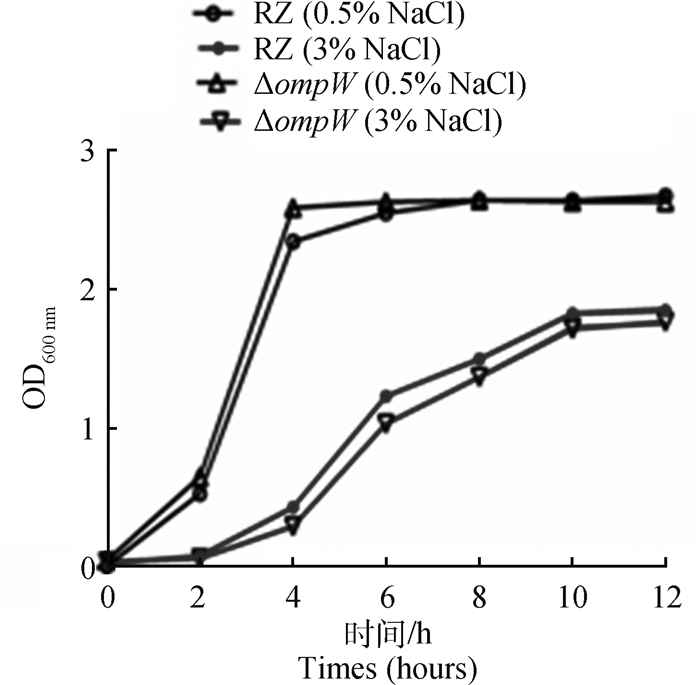
|
图 2 鸭源鸡杆菌RZ和ΔompW在不同NaCl浓度下的生长情况 Fig. 2 Growth of G. anatis RZ and ΔompW in different NaCl concentrations |
用SDS-PAGE及Western blot分析了不同盐度培养基中RZ和ΔompW的外膜蛋白表达情况。鸭源鸡杆菌在3.0%和0.5% NaCl BHI培养时其OmpW相对表达水平分别是0.33和0.23(图 3 B,P>0.05),在高盐度环境下OmpW的表达被上调(图 3A和B)。与RZ相比,在高盐BHI培养时,ΔompW的4种外膜蛋白的表达被上调(图 3 A,虚线箭头所指)。
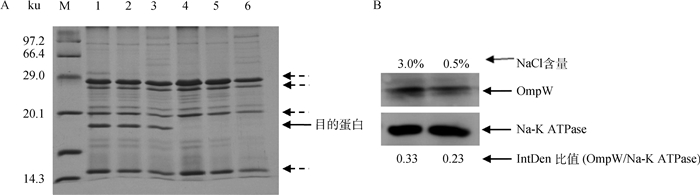
|
A. RZ和ΔompW外膜蛋白的SDS-PAGE分析(泳道1、2、3分别为RZ的外膜蛋白,培养基NaCl含量分别为3.0%,2.0%、0.5%;泳道4、5、6分别为ΔompW外膜蛋白,培养基NaCl含量分别为3.0%、2.0%、0.5%;M.低分子量蛋白质相对分子质量标准);B. RZ OmpW的Western blot(泳道分别为RZ OmpW和内参培养基,NaCl含量分别为3.0%、0.5%;OmpW/Na-K ATPase的IntDen比值对应着图中不同NaCl浓度下OmpW的相对表达) A. SDS-PAGE of outer membrane proteins of RZ (Lane 1, 2, 3. 3.0%, 2.0%, 0.5% NaCl, respectively) and ΔompW (Lane 4, 5, 6. 3.0%, 2.0%, 0.5% NaCl, respectively); Lane M. Protein molecular weight marker (Low); B. Western blot of outer membrane protein OmpW of RZ (The lanes are 3.0%, 0.5% NaCl, respectively). The IntDen ratios of OmpW/CRP corresponding to the lanes of different NaCl concentrations were marked on the top of the figure 图 3 不同盐度下鸭源鸡杆菌RZ和ΔompW OMPs的分析 Fig. 3 Analysis of outer membrane proteins of G. anatis RZ and ΔompW strains under different osmotic stress |
用实时定量PCR方法对正常(0.5% NaCl)和高盐(3.0% NaCl)BHI液体培养基中鸭源鸡杆菌ompW mRNA的表达水平进行分析。根据扩增曲线的Ct值,通过2-△△Ct法计算mRNA的相对表达水平。与低盐BHI(0.5% NaCl)培养下相比,在高盐度BHI中鸭源鸡杆菌ompW mRNA的表达被上调至原来的1.19倍,但差异不显著(P>0.05,图 4)。
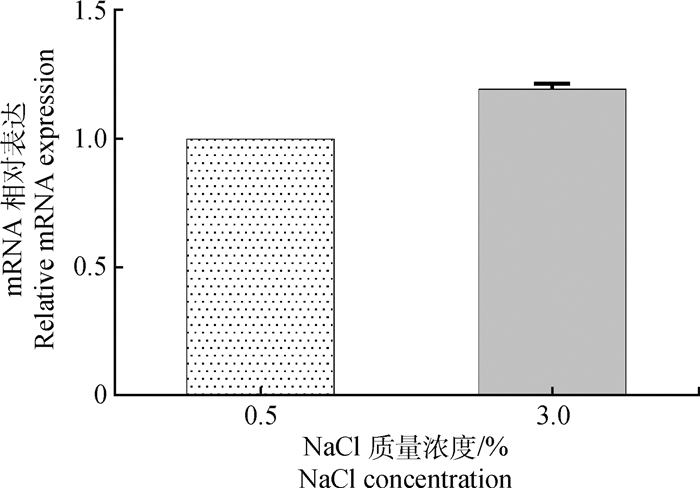
|
图 4 不同盐度下鸭源鸡杆菌ompW mRNA的相对表达水平 Fig. 4 The relative transcription of mRNA of ompW in G. anatis RZ under different osmotis stress |
微生物在长期进化过程中形成了耐受一定应激的机制和能力,使它们具有较强的生存能力。革兰阴性菌表面外膜蛋白不仅有利于其适应复杂环境,而且在与宿主互作中发挥关键作用[27]。外膜是细菌在不同环境中生存的重要保障,能够阻止毒性物质进入细胞。外膜蛋白具有协助小分子的转运、营养摄取、与环境和宿主互作等一系列重要生物学功能[27]。镶嵌在外膜内的孔蛋白是一种跨膜蛋白,通常以β-折叠片构成桶状的非特异性通道,以利于小分子出入细胞;另外,孔蛋白在能量依赖性外排转运中发挥作用[28]。OmpW是小孔蛋白家族成员,形成疏水性通道参与疏水性分子和离子的跨膜转运[29-30]。
外膜蛋白与细菌耐渗透应激密切相关。OmpW导致美人鱼发光杆菌[20]和霍乱弧菌[31]的盐度耐受力下降,OmpW与细菌耐渗透应激相关。与此一致,本研究中,在渗透应激环境下OmpW缺失导致鸭源鸡杆菌生存率下降,且生长性能受到更大影响,这提示OmpW可能与鸭源鸡杆菌的生长和耐渗透应激相关。
细菌外膜蛋白是其适应宿主内环境的关键因素[27],而且外膜蛋白家族包括多种类型外膜蛋白,在抗多类应激反应中发挥作用[29]。大肠杆菌OmpC[32]、克雷伯菌OmpK35[33]和副溶血性弧菌OmpW[20]在不同盐度环境中培养时外膜蛋白的表达被调控。过表达OmpW的大肠杆菌的渗透应激抵抗力提升,而过表达OmpV的大肠杆菌的渗透应激抵抗力下降;OmpF和OmpC是大肠杆菌含量较多的外膜蛋白,在高盐度下,OmpC表达上调,OmpF表达下调;在低盐度下则正好相反[34]。本研究中,在高盐度(3% NaCl)下鸭源鸡杆菌OmpW的表达被上调,且OmpW缺失使鸭源鸡杆菌的盐度耐受性下降,这与上述文献中的研究结果一致。而有趣的是,缺失OmpW ΔompW在高渗透压力中有4种其他类型外膜蛋白的表达被上调(图 3A),这可能是对OmpW缺失的一种生理性补偿调控。
一些相容性溶质可能帮助细菌生存和应对应激,如糖、多元醇、氨基酸和氨基酸衍生物。例如,甜菜碱可被多种细菌用于应对氧化应激[35]。而OmpW是位于细菌外膜的主要蛋白之一,构成运输物质的孔道,在疏水性分子和金属离子转运中发挥重要作用,如OmpW通过转运肉毒碱增强霍乱弧菌的盐耐受能力[31],OmpW通过介导百枯草外排赋予肠道沙门菌耐药性[36]。以往的研究表明细菌在面对高盐应激时会通过排出Na+,摄取K+以适应环境变化。本研究表明,在高盐环境,鸭源鸡杆菌的生存能力和生长性能下降,OmpW缺失株菌受影响更大,因此,我们推测鸭源鸡杆菌OmpW可能通过外排Na+应对渗透应激。OmpW参与鸭源鸡杆菌渗透应激的具体机制有待进一步研究。
4 结论OmpW可能增强了鸭源鸡杆菌的盐耐受性,且鸭源鸡杆菌在高NaCl环境中上调了OmpW的表达。OmpW与鸭源鸡杆菌抗渗透应激相关。
| [1] | CHRISTENSEN H, BISGAARD M, BOJESEN A M, et al. Genetic relationships among avian isolates classified as Pasteurella haemolytica, 'Actinobacillus salpingitidis' or Pasteurella anatis with proposal of Gallibacterium anatis gen. nov., comb. nov. and description of additional genomospecies within Gallibacterium gen. nov[J]. Int J Syst Evol Microbiol, 2003, 53(1): 275–287. DOI: 10.1099/ijs.0.02330-0 |
| [2] | BISGAARD M, KORCZAK B M, BUSSE H J, et al. Classification of the taxon 2 and taxon 3 complex of Bisgaard within Gallibacterium and description of Gallibacterium melopsittaci sp. nov., Gallibacterium trehalosifermentans sp. nov. and Gallibacterium salpingitidis sp. nov[J]. Int J Syst Evol Microbiol, 2009, 59(4): 735–744. DOI: 10.1099/ijs.0.005694-0 |
| [3] | PAUDEL S, LIEBHART D, HESS M, et al. Pathogenesis of Gallibacterium anatis in a natural infection model fulfils Koch's postulates:1. Folliculitis and drop in egg production are the predominant effects in specific pathogen free layers[J]. Avian Pathol, 2014, 43(5): 443–449. DOI: 10.1080/03079457.2014.955782 |
| [4] |
王川庆, 陈陆, 杨霞, 等. 蛋鸡群卡氏杆菌感染情况的初步研究[J]. 河南农业科学, 2008(3): 97–100, 103.
WANG C Q, CHEN L, YANG X, et al. A premilinary study of Gallibacterium anantis infection in laying hens[J]. Journal of Henan Agricultural Sciences, 2008(3): 97–100, 103. DOI: 10.3969/j.issn.1004-3268.2008.03.029 (in Chinese) |
| [5] | SINGH S V, SINGH B R, SINHA D K, et al. Gallibacterium anatis. an emerging pathogen of poultry birds and domiciled birds[J]. J Vet Sci Technol, 2016, 7(3): 1000324. DOI: 10.4172/2157-7579.1000324 |
| [6] |
郭伦涛.鸡杆菌地方分离株耐药性及耐药基因的研究[D].郑州: 河南农业大学, 2010.
GUO L T. Studies on drug resistance and resistant genes of Gallibacterium anatis strains isolated from chickens in different localities[D]. Zhengzhou: Henan Agricultural University, 2010. (in Chinese) http://cdmd.cnki.com.cn/Article/CDMD-10466-2010270671.htm |
| [7] |
彭志锋, 高冬生, 刘红英, 等. 鸭源鸡杆菌整合子及其与耐药性的相关性分析[J]. 畜牧兽医学报, 2016, 47(8): 1676–1681.
PENG Z F, GAO D S, LIU H Y, et al. Gallibacterium anatis integron and its correlation with the drug resistance[J]. Acta Veterinaria et Zootechnica Sinica, 2016, 47(8): 1676–1681. (in Chinese) |
| [8] | BAGER R J, NESTA B, PORS S E, et al. The fimbrial protein FlfA from Gallibacterium anatis is a virulence factor and vaccine candidate[J]. Infect Immun, 2013, 81(6): 1964–1973. DOI: 10.1128/IAI.00059-13 |
| [9] | GARCÍA-GÓMEZ E, VACA S, PÉREZ-MÉNDEZ A, et al. Gallibacterium anatis-secreted metalloproteases degrade chicken IgG[J]. Avian Pathol, 2005, 34(5): 426–429. DOI: 10.1080/03079450500267866 |
| [10] | KRISTENSEN B M, FREES D, BOJESEN A M. Expression and secretion of the RTX-toxin GtxA among members of the genus Gallibacterium[J]. Vet Microbiol, 2011, 153(1-2): 116–123. DOI: 10.1016/j.vetmic.2011.05.019 |
| [11] |
李欢.鸭源鸡杆菌流行株毒素蛋白GtxA表达及其部分生物学特性的研究[D].郑州: 河南农业大学, 2014.
LI H. The expression of protein GtxA and research of some biological characteristics of Gallibacterium anatis[D]. Zhengzhou: Henan Agricultural University, 2014. (in Chinese) http://cdmd.cnki.com.cn/Article/CDMD-10466-1014420759.htm |
| [12] | MONTES-GARCÍA J F, VACA S, VAZQUEZ-CRUZ C, et al. Identification of a Hemagglutinin from Gallibacterium anatis[J]. Curr Microbiol, 2016, 72(4): 450–456. DOI: 10.1007/s00284-015-0969-5 |
| [13] | RAMIREZ-APOLINAR S, GUERRA-INFANTE F M, HARO-CRUZ M D J D, et al. Characterization of a Gallibacterium genomospecies 2 hemagglutinin[J]. J Anim Vet Adv, 2012, 11(4): 556–560. DOI: 10.3923/javaa.2012.556.560 |
| [14] | BAGER R J, PERSSON G, NESTA B, et al. Outer membrane vesicles reflect environmental cues in Gallibacterium anatis[J]. Vet Microbiol, 2013, 167(3-4): 565–572. DOI: 10.1016/j.vetmic.2013.09.005 |
| [15] |
王艳, 王坤芃, 王继洋, 等. 鸭源鸡杆菌外膜蛋白A的原核表达及免疫保护性分析[J]. 中国兽医科学, 2017, 47(10): 1269–1274.
WANG Y, WANG K P, WANG J Y, et al. Prokaryotic expression of Gallibacterium anatis OmpA and immune protection evaluation of the expression product[J]. Chinese Veterinary Science, 2017, 47(10): 1269–1274. (in Chinese) |
| [16] |
彭志锋, 崔丹丹, 杨霞, 等. 鸭源鸡杆菌ompW基因缺失菌株的构建及其药物敏感性分析[J]. 畜牧兽医学报, 2017, 48(7): 1306–1313.
PENG Z F, CUI D D, YANG X, et al. Construction of ompW gene deletion strain of Gallibacterium anatis and its susceotibility to antibacterial agents[J]. Acta Veterinaria et Zootechnica Sinica, 2017, 48(7): 1306–1313. (in Chinese) |
| [17] | HORST R, STANCZAK P, WVTHRICH K. NMR polypeptide backbone conformation of the E. coli outer membrane protein W[J]. Structure, 2014, 22(8): 1204–1209. DOI: 10.1016/j.str.2014.05.016 |
| [18] | XU C X, REN H X, WANG S Y, et al. Proteomic analysis of salt-sensitive outer membrane proteins of Vibrio parahaemolyticus[J]. Res Microbiol, 2004, 155(10): 835–842. DOI: 10.1016/j.resmic.2004.07.001 |
| [19] | MAITI B, RAGHUNATH P, KARUNASAGAR I, et al. Cloning and expression of an outer membrane protein OmpW of Aeromonas hydrophila and study of its distribution in Aeromonas spp.[J]. J Appl Microbiol, 2009, 107(4): 1157–1167. DOI: 10.1111/j.1365-2672.2009.04296.x |
| [20] | WU L N, LIN X M, WANG F P, et al. OmpW and OmpV are required for NaCl regulation in Photobacterium damsela[J]. J Proteome Res, 2006, 5(9): 2250–2257. DOI: 10.1021/pr060046c |
| [21] | YE Y W, LING N, GAO J N, et al. Roles of outer membrane protein W (OmpW) on survival, morphology, and biofilm formation under NaCl stresses in Cronobacter sakazakii[J]. J Dairy Sci, 2018, 101(5): 3844–3850. DOI: 10.3168/jds.2017-13791 |
| [22] | ZHANG X Y, GAO J N, LING N, et al. Short communication:roles of outer membrane protein W on survival, cellular morphology, and biofilm formation of Cronobacter sakazakii in response to oxidative stress[J]. J Dairy Sci, 2019, 102(3): 2017–2021. DOI: 10.3168/jds.2018-14643 |
| [23] | NANDI B, NANDY R K, SARKAR A, et al. Structural features, properties and regulation of the outer-membrane protein W (OmpW) of Vibrio cholerae[J]. Microbiology, 2005, 151(9): 2975–2986. DOI: 10.1099/mic.0.27995-0 |
| [24] |
郑鹿平.我国鸡群鸡杆菌的首次分离鉴定及生物学特性初步研究[D].郑州: 河南农业大学, 2010.
ZHENG L P. The first isolation and identification of Gallibacterium anatis from fowls in china and preliminary research on its biological Characterics[D]. Zhengzhou: Henan Agricultural University, 2010. (in Chinese) http://cdmd.cnki.com.cn/Article/CDMD-10466-2010270665.htm |
| [25] | HUANG J C, WANG X R, CAO Q, et al. ClpP participates in stress tolerance and negatively regulates biofilm formation in Haemophilus parasuis[J]. Vet Microbiol, 2016, 182: 141–149. DOI: 10.1016/j.vetmic.2015.11.020 |
| [26] | KIM S W, CHOI C H, MOON D C, et al. Serum resistance of Acinetobacter baumannii through the binding of factor H to outer membrane proteins[J]. FEMS Microbiol Lett, 2009, 301(2): 224–231. DOI: 10.1111/j.1574-6968.2009.01820.x |
| [27] | LIN J, HUANG S X, ZHANG Q J. Outer membrane proteins:key players for bacterial adaptation in host niches[J]. Microbes Infect, 2002, 4(3): 325–331. DOI: 10.1016/S1286-4579(02)01545-9 |
| [28] | BEKETSKAIA M S, BAY D C, TURNER R J. Outer membrane protein OmpW participates with small multidrug resistance protein member EmrE in quaternary cationic compound efflux[J]. J Bacteriol, 2014, 196(10): 1908–1914. DOI: 10.1128/JB.01483-14 |
| [29] | HATFALUDI T, AL-HASANI K, BOYCE J D, et al. Outer membrane proteins of Pasteurella multocida[J]. Vet Microbiol, 2010, 144(1-2): 1–17. DOI: 10.1016/j.vetmic.2010.01.027 |
| [30] | HONG H, PATEL D R, TAMM L K, et al. The outer membrane protein OmpW forms an eight-stranded β-barrel with a hydrophobic channel[J]. J Biol Chem, 2006, 281(11): 7568–7577. DOI: 10.1074/jbc.M512365200 |
| [31] | FU X P, ZHANG J Y, LI T Y, et al. The outer membrane protein OmpW enhanced V. cholerae growth in hypersaline conditions by transporting carnitine[J]. Front Microbiol, 2018, 8: 2703. DOI: 10.3389/fmicb.2017.02703 |
| [32] | CAI S J, INOUYE M. EnvZ-OmpR interaction and osmoregulation in Escherichia coli[J]. J Biol Chem, 2002, 277(27): 24155–24161. DOI: 10.1074/jbc.M110715200 |
| [33] | HERNÁNDEZ-ALLÉS S, ALBERTI S, ÁLVAREZ D, et al. Porin expression in clinical isolates of Klebsiella pneumoniae[J]. Microbiology, 1999, 145(3): 673–679. DOI: 10.1099/13500872-145-3-673 |
| [34] | JOVANOVICH S B, MARTINELL M, RECORD M T Jr, et al. Rapid response to osmotic upshift by osmoregulated genes in Escherichia coli and Salmonella typhimurium[J]. J Bacteriol, 1988, 170(2): 534–539. DOI: 10.1128/JB.170.2.534-539.1988 |
| [35] | KAPFHAMMER D, KARATAN E, PFLUGHOEFT K J, et al. Role for glycine betaine transport in Vibrio cholerae osmoadaptation and biofilm formation within microbial communities[J]. Appl Environ Microbiol, 2005, 71(7): 3840–3847. DOI: 10.1128/AEM.71.7.3840-3847.2005 |
| [36] | GIL F, IPINZA F, FUENTES J, et al. The ompW (porin) gene mediates methyl viologen (paraquat) efflux in Salmonella enterica serovar typhimurium[J]. Res Microbiol, 2007, 158(6): 529–536. DOI: 10.1016/j.resmic.2007.05.004 |



