2. 省部共建青稞和牦牛种质资源与遗传改良国家重点实验室, 拉萨 850009
2. State Key Laboratory of Hulless Barley and Yak Germplasm Resources and Genetic Improvement, Lhasa 850009, China
黄牛作为优质畜种之一,具有适应能力强、役用性好、饲养范围广等特点,能够提供肉、奶、毛皮等畜产品。我国的黄牛品种资源极其丰富,在农业经济发展中占据重要地位[1]。西藏牛是经过西藏本土改良后的黄牛品种,体型略小于一般黄牛,主要分布于西藏自治区海拔2 300~3 800 m地区,能适应高原地区高海拔、空气稀薄的恶劣气候[2]。其主产区位于西藏雅鲁藏布江中下游、喜马拉雅山东段和三江流域下游地区以及毗邻的草甸草场。三江牛产于我国四川省阿坝藏族羌族自治州,其主产区位于岷山、邛崃山交错的峡谷地带,海拔625~780 m,躯体较长,肉质上佳,免疫力强,是当地人民经过长期选育的优良品种[3]。
脑是由神经元和神经胶质细胞所组成的神经系统控制中心,是所有脊椎动物和大部分无脊椎动物共有的复杂器官。生理方面,脑可以通过分泌化学物质和调节肌肉运动模式的方式调控各个器官协同运作,以应对复杂多变的外部环境[4]。研究表明,哺乳动物神经元组织中circRNA水平较其他组织更丰富。You等[5]通过分析人、小鼠神经细胞株以及ENCODE数据集,发现在所有组织(包括大脑、心、脾、肾以及睾丸等)中大脑的circRNA最多,且有20%的蛋白质编码基因在大脑中形成circRNA。Maass等[6]在对单个人类供体的临床相关组织分析过程中,从大脑皮层中鉴定出339个circRNAs, 其中有141个属于大脑皮层所特有。Xu等[7]探索了不同年龄组恒河猴脑中circRNA的表达变化,共鉴定出17 050个表达良好且稳定的circRNAs,发现circRNA可介导脑老化的生物学调节。Thöken等[8]借助RNA-seq和核酸外切酶富集的方法,在蜜蜂大脑中鉴定获得1 236个circRNAs,发现circRNA的差异表达与蜜蜂在蜂群中的分工存在联系。以上研究成果表明,circRNA在脑功能调控活动中扮演重要角色。
环状RNA(circular RNA, circRNA)是一类由mRNA前体经反向可变剪切形成且大量存在的内源性非编码RNA[9]。较其他非编码RNA而言,circRNA具有环状二级结构,该结构由单链线性转录物经反向剪接以共价键连接形成,特殊的闭合环形结构使circRNA不包含5′端帽子和3′端ploy(A)尾巴,因而对核糖核酸外切酶(RNaseR)具有较强的耐受性,不易被降解[10],常被用作理想的生物标志物,其稳定性约是线性转录物的2.5~5倍[11]。已有的研究表明,circRNA大量存在于真核生物细胞中,长度范围在100~4 000碱基对,呈不均匀分布[12],且丰度较高,具有组织表达特异性和时序表达特异性[13],其序列具有物种保守性。作为一种新型的非编码RNA,circRNA可作为miRNA海绵[14]发挥作用,控制靶向miRNA在细胞质中的表达水平进而解除对mRNA转录物的抑制作用[15],这一作用机制被称为竞争性内源RNA机制。此外,circRNA还能通过干扰可变剪切、结合RNA蛋白等[16]方式发挥其生物调控功能。本研究通过对西藏牛和三江牛的脑组织进行高通量测序,辅以RNaseR酶抗性检测和qRT-PCR验证,旨在从转录本数据中筛选出与低氧适应性相关的差异表达circRNAs,运用生物信息学方法深入分析其相关调控网络及信号通路信息,为后续研究circRNA及其靶基因在西藏牛低氧适应性方面的分子调控机制奠定理论基础。有助于对高原黄牛改良选育工作提供科学的理论支持。
1 材料与方法 1.1 试验动物和样品制备于西藏自治区昌都市类乌齐县(海拔约3 799 m)和四川省阿坝藏族羌族自治州汶川县(海拔约1 325 m),分别选取4.5岁健康、成年雌性西藏牛和三江牛各3头,采集其大脑组织,DEPC水冲洗干净后用锡箔纸包装,迅速置于液氮保存备用。三江牛(Y)和西藏牛(Z)大脑组织样本分别以Y1-DN、Y2-DN、Y3-DN和Z1-DN、Z2-DN、Z3-DN命名。
1.2 西藏牛和三江牛大脑组织RNA提取及质控取适量组织样品于液氮中充分研磨,并迅速转移至含Trizol裂解液的离心管,按照试剂盒说明书提取组织总RNA,-80 ℃保存。利用微量分光光度计NanoDrop 2000测定所提总RNA样本的OD260 mm/OD280 mm值及质量浓度。用Agilent 2100生物分析仪评估RNA质量及完整性。
1.3 西藏牛和三江牛大脑组织cDNA文库构建及测序根据Il-luminaⓇTruSeqTMRNA样品制备步骤,将去除核糖体RNA后的mRNA和非编码RNA(ncRNA)用裂解缓冲液随机打断为小片段,尽可能保留样本中的ncRNA。以此为模板,混合六碱基随机引物、缓冲液、dNTPs、RNase H以及DNA polymerase I合成第二链cDNA。纯化、洗脱经末端修复,添加碱基A和测序接头,采用尿嘧啶-N-糖基化酶(UNG)对cDNA第二链进行降解。利用实时荧光定量PCR系统准确定量cDNA文库,保证文库质量。用Illumina HiSeq 4000平台对质检合格的文库进行双端测序,获得原始测序数据raw reads。
1.4 西藏牛和三江牛大脑组织转录组数据分析为了确保数据质量,需要在信息分析前对原始数据进行质控过滤以减少数据噪音。对得到的原始读段(raw reads)去除含adapter、N比例大于10%以及质量值Q≤10的碱基数占整条read 50%以上的读段,得到可以用于后续信息分析的High Quality Clean Reads(高质量读段)。将每个样品的High Quality Clean Reads与参考基因组(Bos_taurus.ARS-UCD_1.2)进行TopHat比对,提取比对结果中Unmapped reads(未比对上的读段)。截取每条Unmapped Reads的两端(参数为20 bp),通过头尾反向拼接得到Anchors Reads(短序列读段)。将Anchors Reads再次比对到基因组,比对结果提交find_circ软件[17]进行circRNA的筛选和鉴定。对鉴定结果进行过滤,得到高可信度的circRNA。circRNA的表达量由RPM(back-spliced Reads Per Million mapped reads)法计算得到。以西藏牛大脑(Z-DN)和三江牛大脑(Y-DN)作为比较组, 选取|log2FC|>1, P < 0.05为筛选条件,采用DEseq2软件完成差异表达分析。利用比对工具Bowtie2将circRNA两端的anchors reads与基因组进行比对,若两端比对上同一个基因则认定为是circRNA的宿主基因。以Q值≤0.05作为条件,借助R数据包GOseq和KOBAS 3.0软件[18]中普通牛的信息,筛选出显著富集的GO条目和KEGG代谢通路,对宿主基因进行注释。
1.5 circRNA靶向miRNA的预测及互作网络构建通过数据库StarBase[19]找寻差异表达的circRNA与miRNA的靶向关系,运用miRanda和TargetFinder软件对其靶标关系进行预测,获得差异表达circRNA-miRNA靶向调控关系。从结果中选出能够与circRNA靶向结合的miRNA位点,进一步获得circRNA、miRNA、mRNA之间的靶向关系,根据miRNA靶基因的功能注释对此部分的circRNA功能进行阐述。利用Cytoscape v.3.7.1[20]软件绘制circRNA-miRNA-mRNA可视化互作网络图。
1.6 circRNA的验证利用Quantitative Real-time PCR对RNA-seq结果的可靠性进行验证,从鉴定出的差异表达circRNA中随机选取6个进行验证。参照Panda和Gorospe[21]的方法, 运用Primer premier 5.0设计特异性反向引物(表 1),由北京擎科生物科技有限公司进行合成。提取的总RNA分为两份,一份用3 U·mg-1 RNase R(吉赛生物公司,中国)进行消化处理,除去线性RNA,经随机引物反转录后得到相应的cDNA;另一份则用Oligo(dT)23进行反转录得到cDNA,作为后续qPCR的模板,以GAPDH为内参基因,每个样品设置3个重复。试验结果采用2-ΔΔCt法进行计算,运用Origin 9.0和SPSS 18.0软件处理相关数据,完成图表绘制。
|
|
表 1 circRNA及对应的引物序列 Table 1 circRNA and corresponding primer sequences |
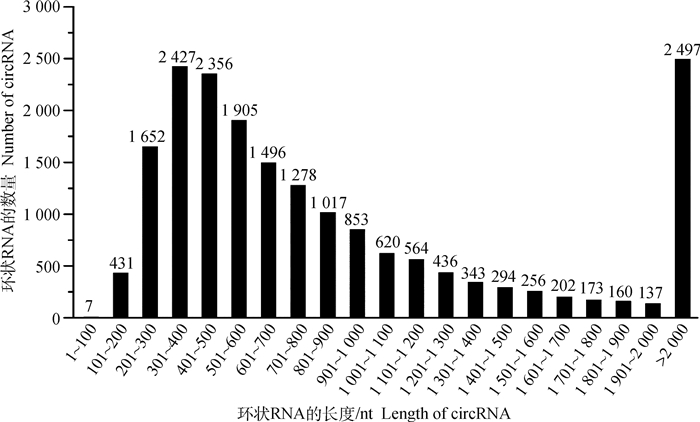
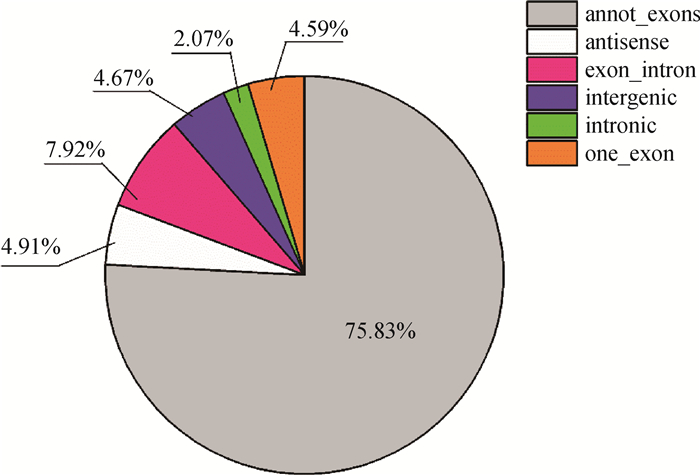
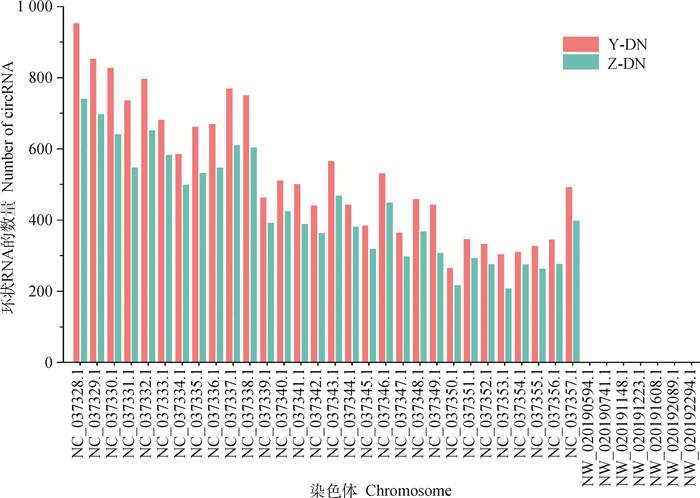
本研究通过选取4.5岁成年健康雌性西藏牛和三江牛共6个样本进行转录组测序,共获得19 130个circRNAs。其raw reads经质控过滤后得到的平均clean reads分别为94 290 294.7条和102 852 192.7条,Q30平均质量百分比分别达到93.26%和94.02%,选取各样本中未比对上核糖体的有效读段比对到参考基因组,得到西藏牛和三江牛的anchors reads平均值分别为11 901 910.7条和18 544 359.3条(表 2),说明测序结果可用于后续分析。由图 1可知,circRNA的长度主要集中于200~1 400 nt之间,平均长度约为2 976 nt,长度区间主要分布于300~400 nt范围内。参考基因组比对结果表明,circRNA可分为annot_exons、antisense、exon_intron、intergenic、intronic、one_exon 6种类型(图 2)。将circRNA比对到基因元件,获得各染色体上circRNA数量的分布情况并绘制分布图(图 3),发现在前30条染色体中,circRNA的数量均高于200,且在相同的单条染色体上三江牛样本circRNA的数量普遍高于西藏牛。
|
|
表 2 Anchors reads与参考基因组比对统计表 Table 2 Anchors reads and reference genome alignment statistics |
|
图 1 circRNA长度分布区间图 Fig. 1 The distribution of circRNA length from sequencing data |
|
图 2 circRNA的类型分布统计图 Fig. 2 Distribution chart of circRNA types |
|
图 3 circRNA在染色体上的数量分布 Fig. 3 Distribution of circRNA on chromosomes |
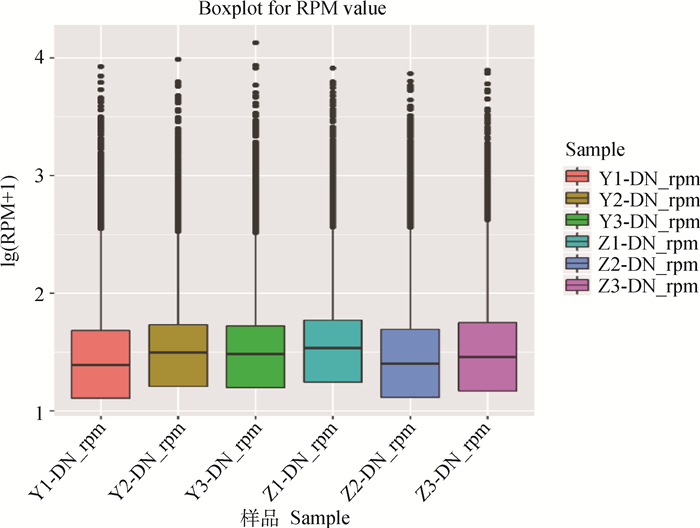
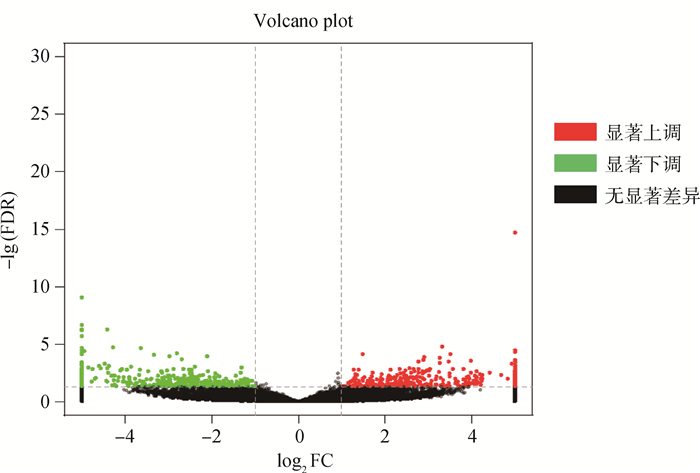
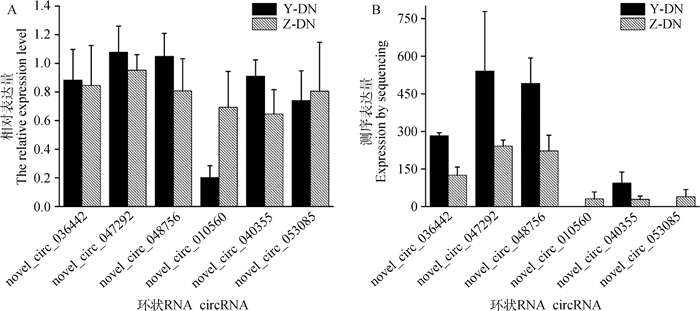
通过RPM法分析circRNA在不同样本中的表达情况,发现其在单个样本中的表达水平均呈现出较高表达趋势(图 4)。运用DEseq2软件进行差异表达分析,共筛选获得858个显著差异表达的circRNAs, 其中包括上调circRNA 394个和下调circRNA 464个(图 5),表 3列举了三江牛和西藏牛大脑组织中前10个显著差异表达的circRNAs信息。从显著差异表达的circRNAs中随机选取6个(novel_ circ_036442、novel_circ_047292、novel_circ_ 048756、novel_circ_010560、novel_circ_040355、novel_circ_053085)进行qRT-PCR验证,以进一步检验其在三江牛和西藏牛大脑组织中的相对表达水平。图 6A展示了6个circRNAs在不同样本中的实时荧光定量结果,这与RNA-seq获得的预测结果(图 6B)趋势相一致,说明本次试验中的测序数据质量真实可靠。
|
图 4 三江牛和西藏牛不同样本中circRNA表达水平箱线图 Fig. 4 Box plot of circRNA expression level in Sanjiang cattle and Zang cattle |
|
横坐标表示差异倍数的对数值,纵坐标表示校正后P值的负log10转化值 The X axis represent the logarithmic value of Fold Change, the Y axis represent the negative log10 conversion value of the corrected P-value 图 5 circRNA的显著差异表达谱 Fig. 5 Significant differential expression profile of circRNA |
|
|
表 3 差异显著前10的circRNAs Table 3 Top 10 signifcant regulated circRNAs |
|
图 6 三江牛和西藏牛大脑组织间6个随机差异表达circRNA的验证(A)和测序结果(B) Fig. 6 Verification (A) and sequencing results (B) of 6 random differentially expressed circRNAs in the brains between Sanjiang cattle and Zang cattle |
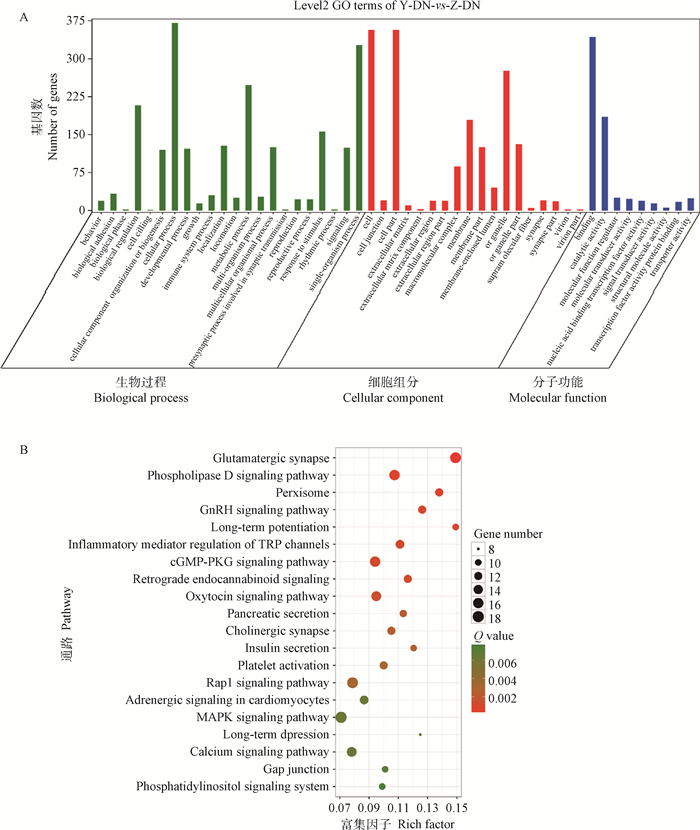
circRNA由蛋白质编码基因的外显子通过反向剪接产生。因此,对其宿主基因进行注释在一定程度上可以诠释circRNA的功能。GO数据库比对结果显示,显著差异表达circRNA的宿主基因可注释到49个功能亚类中,涉及生物学过程(biological process)、细胞组分(cellular component)以及分子功能(molecular function)3个大类(图 7A)。其中生物学过程主要富集在细胞过程(cellular process)、单一生物过程(single-organism process)、代谢过程(metabolic process)、生物调节(biological regulation)、应激反应(response to stimulus)等5个GO条目, 且分别有30、22、19、2个宿主基因注释到免疫系统过程(immune system process)、再生(reproduction)、行为(behavior)和突触前过程涉及突触传递(presynaptic process involved in synaptic transmission);细胞组分主要富集在细胞(cell)、细胞组分(cell part)、细胞器(organelle)、膜(membrane)等条目,其中有87个宿主基因注释到大分子复合物(macromolecular complex);分子功能方面,宿主基因主要分布在粘合物(binding)、催化活性(catalytic activity)、分子功能调节(molecular function regulator)、转运活动(transporter activity)以及分子换能器活动(molecular transducer activity)。上述结果表明,circRNA可能通过调节细胞中大分子复合物的活动,在西藏牛脑功能的代谢过程、生物调节和应激反应等生物学过程中发挥作用。
|
图 7 差异表达circRNA宿主基因GO分析(A)和KEGG通路富集(B) Fig. 7 GO (A) and KEGG pathway enrichment (B) analysis of the differentially expressed circRNA host genes |
差异表达circRNA宿主基因的KEGG分析结果共注释到31个显著富集的信号通路(P < 0.05),气泡图(图 7B)显示了KEGG PATHWAY中前20的KEGG富集情况。结果表明,circRNA的宿主基因在MAPK信号通路(MAPK signaling pathway,ko04010)的富集程度最高,其次是谷氨酸能突触(glutamatergic synapse,ko04724)、Rap1信号通路(Rap1 signaling pathway,ko04015)、磷脂酶D信号通路(phospholipase D signaling pathway,ko04072)以及cGMP-PKG信号通路(cGMP-PKG signaling pathway,ko04022)等。
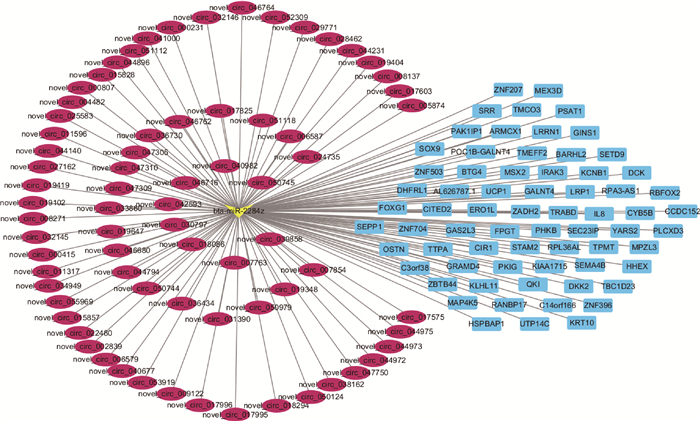
2.4 miRNA的靶点预测及circRNA-miRNA-mRNA网络分析circRNA作为高效的竞争性内源RNA,富含miRNA结合位点,且能够通过miRNA海绵效应吸附miRNA进而解除其对靶向mRNA的抑制作用,提高靶基因表达水平[22]。本研究采用miRanda(v3.3)和TargetScan(Version:7.1)软件借助Linux平台预测circRNA与miRNA的结合位点及相关靶基因。在350个上调和364个下调差异表达circRNAs中分别筛选获得492和508个miRNAs靶点,其中miRNA靶点数最多的是novel_circ_017825(235个)和novel_circ_046762(231个),说明它们可能在西藏牛脑功能调控方面具有重要作用。图 8A为根据上述circRNA/miRNA的靶向关系绘制的差异表达circRNA-miRNA全景调控网络。其次,将图中的部分调控网络进行放大,图 8B展示了5个(在novel_circ_036442中未预测到靶向miRNA)RT-qPCR验证的circRNAs及其作用的miRNAs间的靶向关系。miRNA靶点的统计结果表明,bta-miR-2284z能够向上结合最多的circRNA(73个),并向下与诸如IL-8(白细胞介素8)、UCP1(解偶联蛋白1)、HHEX(造血表达同源框)、YARS2(酪氨酰-tRNA合成酶2)等68个mRNAs存在靶向关系(图 9)。由此推测,这些circRNAs可能通过miRNA海绵效应吸附bta-miR-2284z,间接影响西藏牛脑组织中相关靶向mRNA的表达水平。

|
A.显著差异表达circRNA-miRNA的全景调控网络;B. qRT-PCR验证的circRNA-miRNA靶向关系图。红色节点表示上调的circRNAs,黄色节点表示下调的circRNAs,蓝色节点表示miRNAs,节点间的连线表示种子序列配对的相互作用 A. Panoramic regulatory network of significantly differentially expressed circRNA-miRNA; B. circRNA-miRNA targeting diagram verified by qRT-PCR; The red nodes represent up-regulated circRNAs, the yellow nodes represent down-regulated circRNAs, and the blue nodes represent miRNAs, the lines between nodes represent interaction of seed sequence pairing 图 8 差异表达circRNA-miRNA调控网络 Fig. 8 Differentially expressed circRNA-miRNA regulatory network |
|
椭圆表示circRNAs,箭头表示miRNA,矩形表示miRNA靶基因,节点间的连线表示序列间的配对关系 The ellipses represent circRNAs, the arrow represents miRNA, and the rectangles represent target genes of miRNA, the lines between nodes represent the pairing relationship between sequences 图 9 差异表达circRNA-bta-miR-2284z-mRNA调控网络 Fig. 9 Regulatory network of differentially expressed circRNA-bta-miR-2284z-mRNA |
circRNA因其特殊的拓扑结构,对核酸外切酶RNaseR具有很强的抗性。如图 10所示,经RNaseR消化后,较对照组RNaseR (-),处理组RNaseR (+)中仍能够检测到circRNA,表达水平比前者明显升高,个别circRNA呈现倍数增长。琼脂糖凝胶电泳检测和Sanger测序结果表明所获得的circRNA是环状转录本。
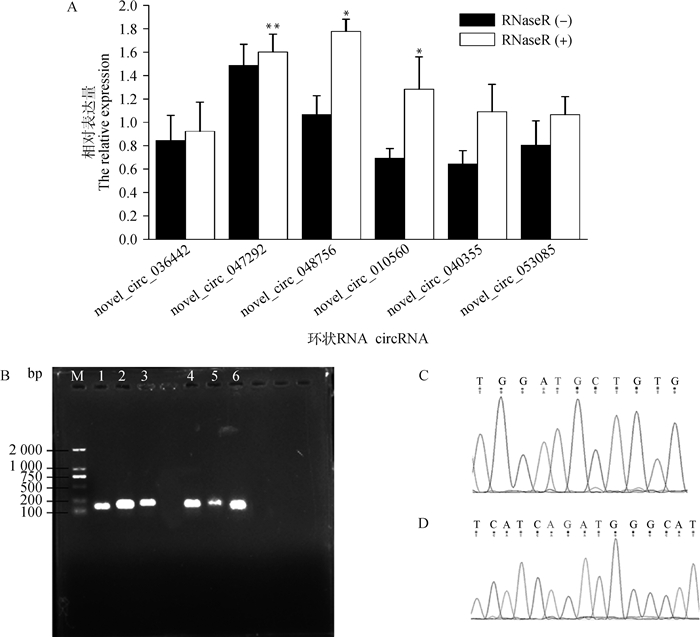
|
A. RNaseR酶抗性验证结果:数据表示为“平均值± SEM”,**. P<0.01,*. P<0.05;B. qRT-PCR产物的电泳图:M. DNA相对分子质量标准,1~6. novel_circ_036442、novel_circ_047292、novel_circ_048756、novel_circ_010560、novel_circ_040355、novel_circ_053085;C、D. Sanger测序证实的具有代表性的两例qPCR产物,C表示novel_circ_036442,D表示novel_circ_047292 A. The result of RNaseR enzyme resistance verification: The data are presented as the "mean ± SEM", **.P < 0.01, *. P < 0.05; B. Electrophoresis of qRT-PCR product: M. DL2000 marker; 1-6. novel_circ_036442, novel_circ_047292, novel_circ_048756, novel_circ_010560, novel_circ_040355, novel_circ_053085; C, D. Two representative examples of qPCR products confirmed by Sanger sequencing, C is novel_circ_036442, D is novel_circ_047292 图 10 circRNA的RNaseR酶抗性和Sanger测序 Fig. 10 RNaseR enzyme resistance and Sanger sequencing for circRNA |
随着近些年生物信息学的快速发展和RNA-seq测序技术的不断成熟,针对circRNA特性设计的软件工具层出不穷[23]。传统的RNA测序程序依赖于5′末端或3′聚(A)尾的纯化步骤,导致闭环结构的circRNA在最初的标准化处理过程中常作为副产物被去除[24],而诸如CIRI2、find_circ、KNIFE等circRNA算法程序却能有效的适用这一点。目前,已经在小鼠[25]、人[26]、猴子[27]、猪[28]、蜜蜂[29]、牛[30]、羊[31]等物种中发现了大量的circRNAs, 并挖掘出许多在影响mRNA表达水平方面具有重要调控功能的circRNA。本研究采用高通量测序技术分别对三江牛和西藏牛大脑组织进行建库、测序,利用find_circ软件从6个样本中共检测出19 130个circRNAs。这与Gao等[32]通过去除rRNA和线性RNA构建文库的方法从成年秦川牛睾丸组织中鉴定出的circRNA数量相当,说明在circRNA预测前期,采用非去线性RNA建库的方法具有可行性。此外,circRNA的长度分布统计情况显示,绝大多数circRNA的长度均在400 nt以上,这与前人在树鼩[33]脑组织中的研究发现相一致。说明circRNA的长度分布特征可能在哺乳动物脑组织中具有普遍性。其次,以|log2FC|>1, P < 0.05作为筛选条件,在西藏牛和三江牛中筛选出显著差异表达的circRNA共858个,其中394个上调,464个下调。利用qRT-PCR验证novel_circ_036442、novel_circ_047292、novel_circ_048756、novel_circ_010560、novel_circ_040355、novel_circ_053085等6个随机选择的circRNAs,表达情况与RNA-seq预测的表达趋势一致。说明本次测序数据真实可靠。RNaseR酶抗性检测结果显示,circRNA对RNaseR酶的消化作用具有耐受性,且部分circRNA的表达水平相比对照组呈现倍数增长。结合琼脂糖凝胶电泳和Sanger测序结果证明所获得的circRNA是环状转录本。
为了进一步探索circRNA与低氧适应相关性,对其宿主基因进行功能注释和KEGG信号通路分析。GO注释结果表明,这些显著差异表达circRNA的宿主基因被注释到49个功能亚类。其中,生物学过程主要涉及到细胞过程、单一生物过程、代谢过程、生物调节、应激反应等,此外,还包括免疫系统、再生和突触传递。前人的研究表明,大脑神经组织中表达的circRNA很大一部分来自突触基因,能够参与诱导神经元的应激反应[34]。突触是脑组织的关键功能元素。在极度缺氧环境下,兴奋性突触传递会被快速抑制造成脑部缺氧性损伤。本研究中,分别有2个和30个circRNAs注释到突触传递和免疫系统过程,156个circRNAs与应激反应相关,推测这些circRNAs可能通过介导脑组织中相关神经元的突触传递过程影响西藏牛在高原低氧环境下的免疫调节功能和应激反应。此外,在KEGG通路分析中,共获得31个显著富集的信号通路(P < 0.05),主要涉及MAPK信号通路、谷氨酸能突触、Rap1信号通路、磷脂酶D信号通路以及cGMP-PKG信号通路,其中还包括钙信号通路、血小板活化等。研究表明,MAPK信号通路[35-38]、谷氨酸能突触[39]、Rap1信号通路[40]与脑损伤修复、细胞免疫、Ca+通道以及细胞迁移密切相关。本研究统计显示,差异表达circRNA的宿主基因分别有18和17个注释到MAPK信号通路、谷氨酸能突触,17个注释到Rap1信号通路。由此推断,上述circRNAs可能通过MAPK信号通路、谷氨酸能突触、Rap1信号通路在西藏牛脑部缺氧性损伤修复、细胞免疫、Ca+通道及细胞迁移等生物学过程发挥重要调控作用。
研究表明,circRNA可作为竞争性内源RNA,通过miRNA反应元件竞争性结合共同的miRNA。例如,Lei等[41]通过研究正常脑组织和胶质瘤脑组织,揭示了hsa_circ_0076248作用海绵体miR-181a在胶质瘤生长和侵袭中的调节机制。Zhang等[42]通过RNA-seq分析了肿瘤组织和邻近正常胃黏膜组织,发现circNRIP1-miR-149-5p-AKT1/Mtor这一新的调控路径,证明了circNRIP1可靶向调控miR-149-5p,进而影响AKT1的表达水平,并最终在胃癌中作为肿瘤启动子。本研究通过构建circRNA-miRNA互作网络,从350个上调和364个下调的差异表达circRNAs中分别预测得到492和508个miRNAs靶点,其中miRNA靶点数最多的是novel_circ_017825和novel_circ_046762,说明circRNA具有靶向结合多条不同miRNAs的能力。其次,circRNA-bta-miR-2284z-mRNA调控网络显示,bta-miR-2284z能够靶向结合最多的circRNA,并靶向映射IL-8(白细胞介素8)、UCP1(解偶联蛋白1)、HHEX(造血表达同源框)、YARS2(酪氨酰-tRNA合成酶2)等68个mRNAs。查询miRBase数据库[43]发现,bta-miR-2284z与bta-miR-2284具有较高的同源性,且同属于bta-miR-2284家族[44]。已有的研究表明,bta-miR-2284能够与TLR基因协同作用产生促炎细胞因子,并进一步调节非特异性免疫应答[45]。由此推测,上述差异表达的circRNAs可能通过竞争性结合bta-miR-2284z的方式,在调控西藏牛脑组织细胞的非特异性免疫应答过程中发挥重要作用。但相关转录因子的具体作用机制目前尚不清楚,还需要后续研究证明。
4 结论本研究采用高通量测序技术获得了三江牛和西藏牛大脑组织中circRNA的表达谱和差异表达信息。在样本中检测到丰富的circRNAs,并对其基因类型、长度分布及基因组特征进行统计。circRNA宿主基因的功能分析表明,所筛选出的858个circRNA主要与MAPK信号通路、谷氨酸能突触、Rap1信号通路、磷脂酶D信号通路以及cGMP- PKG信号通路等有关。利用生物信息学方法预测circRNA、miRNA及其靶向mRNA间的联系,并初步构建circRNA-miRNA-mRNA网络互作模型。为后续进一步探索circRNA参与西藏牛低氧适应性的生物学过程和分子功能,研究circRNA在大型哺乳动物低氧适应性方面的调控机制提供数据支持。
| [1] |
黄兴, 柴志欣, 信金伟, 等. 西藏牛亚科部分品种线粒体DNA遗传多样性研究[J]. 西南民族大学学报:自然科学版, 2019, 45(2): 117–124.
HUANG X, CHAI Z X, XIN J W, et al. Genetic diversity of mitochondrial DNA of some subfamilies of cattle in Tibet[J]. Journal of Southwest Minzu University:Natural Science Edition, 2019, 45(2): 117–124. (in Chinese) |
| [2] |
王莉, 旦增洛桑, 马金英, 等. 娟姗牛杂交改良西藏本地黄牛的效果调查[J]. 中国奶牛, 2015(2): 13–15.
WANG L, DANZENG L S, MA J Y, et al. Hybrid improvement of jersey cattle crossbreeding with Tibet local yellow cattle[J]. China Dairy Cattle, 2015(2): 13–15. DOI: 10.3969/j.issn.1004-4264.2015.02.004 (in Chinese) |
| [3] |
郭琳, 柴志欣, 钟金城, 等. 三江黄牛DKK1、DKK4和MyoG基因遗传多态性分析[J]. 家畜生态学报, 2018, 39(6): 16–20.
GUO L, CHAI Z X, ZHONG J C, et al. Genetic diversity of DKK1, DKK4 and MyoG in Sanjiang cattle[J]. Acta Ecologae Animalis Domastici, 2018, 39(6): 16–20. DOI: 10.3969/j.issn.1673-1182.2018.06.004 (in Chinese) |
| [4] | MA Q F, LI L Z, YU B X, et al. Circular RNA profiling of neutrophil transcriptome provides insights into asymptomatic Moyamoya disease[J]. Brain Res, 2019, 1719: 104–112. DOI: 10.1016/j.brainres.2019.05.033 |
| [5] | YOU X T, VLATKOVIC I, BABIC A, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity[J]. Nat Neurosci, 2015, 18(4): 603–610. DOI: 10.1038/nn.3975 |
| [6] | MAASS P G, GLAŽR P, MEMCZAK S, et al. A map of human circular RNAs in clinically relevant tissues[J]. J Mol Med (Berl), 2017, 95(11): 1179–1189. DOI: 10.1007/s00109-017-1582-9 |
| [7] | XU K Y, CHEN D, WANG Z B, et al. Annotation and functional clustering of circRNA expression in rhesus macaque brain during aging[J]. Cell Discov, 2018, 4: 48. |
| [8] | THÖKEN C, THAMM M, ERBACHER C, et al. Sequence and structural properties of circular RNAs in the brain of nurse and forager honeybees (Apis mellifera)[J]. BMC Genomics, 2019, 20(1): 88. DOI: 10.1186/s12864-018-5402-6 |
| [9] | LI X, YANG L, CHEN L L. The biogenesis, functions, and challenges of circular RNAs[J]. Mol Cell, 2018, 71(3): 428–442. DOI: 10.1016/j.molcel.2018.06.034 |
| [10] |
张进威, 龙科任, 王讯, 等. 环状RNA研究进展[J]. 畜牧兽医学报, 2016, 47(11): 2151–2158.
ZHANG J W, LONG K R, WANG X, et al. The research advance of circular RNA[J]. Acta Veterinaria et Zootechnica Sinica, 2016, 47(11): 2151–2158. DOI: 10.11843/j.issn.0366-6964.2016.11.001 (in Chinese) |
| [11] | ENUKA Y, LAURIOLA M, FELDMAN M E, et al. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor[J]. Nucleic Acids Res, 2016, 44(3): 1370–1383. DOI: 10.1093/nar/gkv1367 |
| [12] | LASDA E, PARKER R. Circular RNAs:diversity of form and function[J]. RNA, 2014, 20(12): 1829–1842. DOI: 10.1261/rna.047126.114 |
| [13] |
吴艳, 潘爱銮, 杜金平, 等. 环状RNA及其在畜禽中的研究进展[J]. 西北农业学报, 2018, 27(3): 301–305.
WU Y, PAN A L, DU J P, et al. Research advance of circular RNA in livestock and poultry[J]. Acta Agriculturae Boreali-occidentalis Sinica, 2018, 27(3): 301–305. (in Chinese) |
| [14] | HANSEN T B, JENSEN T I, CLAUSEN B H, et al. Natural RNA circles function as efficient microRNA sponges[J]. Nature, 2013, 495(7441): 384–388. DOI: 10.1038/nature11993 |
| [15] | QU S B, YANG X S, LI X L, et al. Circular RNA:a new star of noncoding RNAs[J]. Cancer Lett, 2015, 365(2): 141–148. DOI: 10.1016/j.canlet.2015.06.003 |
| [16] | HANSEN T B, WIKLUND E D, BRAMSEN J B, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA[J]. EMBO J, 2011, 30(21): 4414–4422. DOI: 10.1038/emboj.2011.359 |
| [17] | MEMCZAK S, JENS M, ELEFSINIOTI A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency[J]. Nature, 2013, 495(7441): 333–338. DOI: 10.1038/nature11928 |
| [18] | XIE C, MAO X Z, HUANG J J, et al. KOBAS 2.0:a web server for annotation and identification of enriched pathways and diseases[J]. Nucleic Acids Res, 2011, 39(S2): W316–W322. |
| [19] | LI J H, LIU S, ZHOU H, et al. StarBase v2.0:decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data[J]. Nucleic Acids Res, 2014, 41(D1): D92–D97. |
| [20] | SHANNON P, MARKIEL A, OZIER O, et al. Cytoscape:a software environment for integrated models of biomolecular interaction networks[J]. Genome Res, 2003, 13(11): 2498–2504. DOI: 10.1101/gr.1239303 |
| [21] | PANDA A C, GOROSPE M. Detection and analysis of circular RNAs by RT-PCR[J]. Bio Protoc, 2018, 8(6): e2775. |
| [22] | CHEN L L. The biogenesis and emerging roles of circular RNAs[J]. Nat Rev Mol Cell Biol, 2016, 17(4): 205–211. DOI: 10.1038/nrm.2015.32 |
| [23] | HANSEN T B, VENØ M T, DAMGAARD C K, et al. Comparison of circular RNA prediction tools[J]. Nucleic Acids Res, 2016, 44(6): e58. DOI: 10.1093/nar/gkv1458 |
| [24] | JECK W R, SHARPLESS N E. Detecting and characterizing circular RNAs[J]. Nat Biotechnol, 2014, 32(5): 453–461. DOI: 10.1038/nbt.2890 |
| [25] | JIANG L, LI H J, FAN Z M, et al. Circular RNA expression profiles in neonatal rats following hypoxic-ischemic brain damage[J]. Int J Mol Med, 2019, 43(4): 1699–1708. |
| [26] | ZHANG M J, JIA L F, ZHENG Y F. circRNA expression profiles in human bone marrow stem cells undergoing osteoblast differentiation[J]. Stem Cell Rev Rep, 2019, 15(1): 126–138. DOI: 10.1007/s12015-018-9841-x |
| [27] | ABDELMOHSEN K, PANDA A C, DE S, et al. Circular RNAs in monkey muscle:age-dependent changes[J]. Aging (Albany NY), 2015, 7(11): 903. |
| [28] | LI A, HUANG W, ZHANG X, et al. Identification and characterization of CircRNAs of two pig breeds as a new biomarker in metabolism-related diseases[J]. Cell Physiol Biochem, 2018, 47(6): 2458–2470. DOI: 10.1159/000491619 |
| [29] | CHEN X, SHI W, CHEN C. Differential circular RNAs expression in ovary during oviposition in honey bees[J]. Genomics, 2019, 111(4): 598–606. DOI: 10.1016/j.ygeno.2018.03.015 |
| [30] | HUANG J P, ZHAO J H, ZHENG Q Z, et al. Characterization of circular RNAs in Chinese buffalo (Bubalus bubalis) adipose tissue:a focus on circular RNAs involved in fat deposition[J]. Animals (Basel), 2019, 9(7): 403. DOI: 10.3390/ani9070403 |
| [31] |
邹双霞, 金澄艳, 鲍建军, 等. 感染大肠杆菌F17湖羊羔羊脾脏中差异circRNA分析[J]. 中国农业科学, 2019, 52(6): 1090–1101.
ZOU S X, JIN C Y, BAO J J, et al. Differential circRNA analysis in the spleen of Hu-sheep lambs infected with F17 Escherichia coli[J]. Scientia Agricultura Sinica, 2019, 52(6): 1090–1101. (in Chinese) |
| [32] | GAO Y, WU M L, FAN Y Z, et al. Identification and characterization of circular RNAs in Qinchuan cattle testis[J]. R Soc Open Sci, 2018, 5(7): 180413. DOI: 10.1098/rsos.180413 |
| [33] | LU C X, SUN X M, LI N, et al. CircRNAs in the tree shrew (Tupaia belangeri) brain during postnatal development and aging[J]. Aging (Albany NY), 2018, 10(4): 833–852. |
| [34] | VAN ROSSUM D, VERHEIJEN B M, PASTERKAMP R J. Circular RNAs:novel regulators of neuronal development[J]. Front Mol Neurosci, 2016, 9: 74. |
| [35] | CHEN J, AGUILERA G. Vasopressin protects hippocampal neurones in culture against nutrient deprivation or glutamate-induced apoptosis[J]. J Neuroendocrinol, 2010, 22(10): 1072–1081. DOI: 10.1111/j.1365-2826.2010.02054.x |
| [36] | KIM S W, LIM C M, KIM J B, et al. Extracellular HMGB1 released by NMDA treatment confers neuronal apoptosis via RAGE-p38 MAPK/ERK signaling pathway[J]. Neurotox Res, 2011, 20(2): 159–169. DOI: 10.1007/s12640-010-9231-x |
| [37] | SARAAV I, SINGH S, SHARMA S. Outcome of Mycobacterium tuberculosis and Toll-like receptor interaction:immune response or immune evasion?[J]. Immunol Cell Biol, 2014, 92(9): 741–746. DOI: 10.1038/icb.2014.52 |
| [38] | TAO L, LI D, LIU H X, et al. Neuroprotective effects of metformin on traumatic brain injury in rats associated with NF-κB and MAPK signaling pathway[J]. Brain Res Bull, 2018, 140: 154–161. DOI: 10.1016/j.brainresbull.2018.04.008 |
| [39] |
李晶, 杜永平, 张月萍. 缺氧对谷氨酸能和GABA能突触传递的影响[J]. 中国病理生理杂志, 2013, 29(2): 371–375.
LI J, DU Y P, ZHANG Y P. Effects of hypoxia on glutamatergic and GABAergic synaptic transmission[J]. Chinese Journal of Pathophysiology, 2013, 29(2): 371–375. DOI: 10.3969/j.issn.1000-4718.2013.02.033 (in Chinese) |
| [40] | SHAH S, BROCK E J, JI K, et al. Ras and Rap1:a tale of two GTPases[J]. Semin Cancer Biol, 2019, 54: 29–39. DOI: 10.1016/j.semcancer.2018.03.005 |
| [41] | LEI B X, HUANG Y T, ZHOU Z W, et al. Circular RNA hsa_circ_0076248 promotes oncogenesis of glioma by sponging miR-181a to modulate SIRT1 expression[J]. J Cell Biochem, 2019, 120(4): 6698–6708. DOI: 10.1002/jcb.27966 |
| [42] | ZHANG X, WANG S, WANG H X, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway[J]. Mol Cancer, 2019, 18(1): 20. DOI: 10.1186/s12943-018-0935-5 |
| [43] | GRIFFITHS-JONES S, SAINI H K, VAN DONGEN S, et al. miRBase:tools for microRNA genomics[J]. Nucleic Acids Res, 2008, 36(S1): D154–D158. |
| [44] | JIN W W, IBEAGHA-AWEMU E M, LIANG G X, et al. Transcriptome microRNA profiling of bovine mammary epithelial cells challenged with Escherichia coli or Staphylococcus aureus bacteria reveals pathogen directed microRNA expression profiles[J]. BMC Genomics, 2014, 15: 181. DOI: 10.1186/1471-2164-15-181 |
| [45] | DAS K, GARNICA O, DHANDAYUTHAPANI S. Modulation of host miRNAs by intracellular bacterial pathogens[J]. Front Cell Infect Microbiol, 2016, 6: 79. |



