2. State Key Laboratory of Brain and Cognitive Science, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China ;
3. State Key Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences Chinese Academy of Medical Sciences & School of Basic Medicine Peking Union Medical College, Beijing 100005, China
Objective This study is aimed at observing the role of long noncoding RNAs (lncRNAs) in the pathogenesis of abdominal aortic aneurysm (AAA).
Methods LncRNA and mRNA expression signatures of AAA tissues and normal abdominal aortic tissues (NT) were analyzed by microarray and further verified by Real-time quantitative reverse-transcription PCR (qRT-PCR). The lncRNAs-mRNAs targeting relationships were identified using computational analysis. The effect of lnc-ARG on 5-lipoxygenase (ALOX5) expression was tested in HeLa cells.
Results Differential expressions of 3,688 lncRNAs and 3,007 mRNAs were identified between AAA and NT tissues. Moreover, 1,284 differentially expressed long intergenic noncoding RNAs and 206 differentially expressed enhancer-like lncRNAs adjacent to protein-coding genes were discerned by bioinformatics analysis. Some differentially expressed lncRNAs and mRNAs between AAA and normal tissue samples were further verified using qRT-PCR. A co-expression network of coding and noncoding genes was constructed based on the correlation analysis between the differentially expressed lncRNAs and mRNAs. In addition, the lnc-ARG located within the upstream of ALOX5 was sorted as a noncoding transcript by analyzing the protein-coding potential using computational analysis. Furthermore, we found that lnc-ARG can decrease the mRNA level of ALOX5 and reactive oxygen species production in HeLa cells.
Conclusion This study revealed new lncRNA candidates are related to the pathogenesis of AAA.
Abdominal aortic aneurysm (AAA) is defined as a maximum infrarenal abdominal aortic aneurysm with a diameter of 3.0 cm or more and was characterized by destruction of elastin and collagen in the aortic media and adventitia[1]. The pathological features of AAA are characterized by extracellular matrix remodeling,loss of integrity of the arterial wall,vascular smooth muscle cell (VSMC) apoptosis,infiltration of inflammatory cells,and oxidative stress[2]. An imbalance between extracellular matrix synthesis and degradation favoring catabolic processes is believed to be critical in AAA pathogenesis. Matrix metalloproteinases (MMPs) have been shown to be the most abundant proteases expressed in AAA wall[3]. The activation of MMPs is tightly regulated by tissue inhibitors of metalloproteinases (TIMPs),and mRNA levels of TIMPs were decreased in AAA tissue[4-5]. Other proteases were also reported to contribute to the initiation and progression of AAA,such as cysteine proteases[6-7]. Previous studies suggest that apoptosis and depletion of VSMCs make an important contribution to AAA by eliminating a cell population capable of directing connective tissue repair. Chronic inflammation of the aortic wall plays an important role in the pathogenesis of AAA. Studies of human AAA tissues have shown extensive inflammatory infiltrates containing macrophages,lymphocytes,and mast cells in both the media and adventitia,and increasing aneurysm diameter was associated with a higher density of inflammatory cells in the adventitia[8]. Some mediators ,including several proteases,proinflammatory cytokines,growth factors,and chemokines synthesized and released by infiltrated inflammatory cells,induce adventitial inflammation,apoptosis of VSMCs,activation of MMPs,and neovascularization in the arterial wall[2]. Reactive oxygen species (ROS) production was increased in the aneurysm wall compared with the normal aorta and adjacent non-aneurysmal aortic wall[9]. All of the above mentioned,including infiltrated inflammatory cells,proinflammatory cytokines,mechanical stretch,growth factors,and lipid mediators,might upregulate NADPH oxidase in resident vascular cells,resulting in an increase in the production of ROS and lipid peroxidation products[10]. Overexpressed ROS and nitric oxide increased the expression of MMPs through the activation of nuclear factor-kappa B and induced apoptosis of VSMCs in the aneurysm wall[11].
Noncoding RNAs can be arbitrarily divided into small noncoding RNAs and long noncoding RNAs (lncRNAs) according to their length. LncRNAs are defined as noncoding RNAs longer than 200 nucleotides in length,transcribed by RNA polymerase II (RNA pol II),spliced,capped,and polyadenylated,lacking significant open reading frames,and cannot encode proteins[12]. Collective evidence indicates that lncRNAs have comprehensive biological functions through various mechanisms and are correlated with both normal development progress and diseases,such as cancer[13]. Studies on the involvement of lncRNAs in cardiology and vascular biology are scarce to date. However,some promising studies indicate that some lncRNAs have been found to be differentially regulated in the developing or diseased heart and can provide a strong indication for their involvement in cardiac (patho)physiology[14]. Previous studies showed that microRNAs have been confirmed to be involved in AAA development[15-18]. However,the role of lncRNAs in AAA remains to be clarified.
In this study,we analyzed the expression profiles of lncRNAs and mRNAs in human AAA tissues and normal tissues collected from different subjects by microarray. Bioinformatics analysis was performed to predict the target mRNAs of lncRNAs by combining differentially expressed mRNAs and lncRNAs. Among them,we found that lncRNA-ARG inhibited the mRNA level of 5-lipoxygenase (ALOX5),a key enzyme involved in the synthesis of proinflammatory leukotrienes (LTs),and reduced ROS production.
MATERIALS AND METHODS Patients and Tissue SamplesThree AAA patients and three control subjects (aged 55-65 years) were recruited for microarray analysis of lncRNAs. The AAA tissues were acquired by surgery and normal abdominal aortic tissues (NT) were obtained from subjects with physical traumas. AAA tissues and histologically matched NT from each subject were snap-frozen in liquid nitrogen immediately after resection. All participating subjects provided a written informed consent. The study was approved by the Ethics Committee of the Medical Faculty of Beijing Anzhen Hospital,Capital Medical University,Beijing,China and was conducted in accordance with the World Medical Association Declaration of Helsinki.
RNA ExtractionTotal RNA was extracted from AAA and matched normal tissues using Trizol reagent (Invitrogen,Carlsbad,CA,USA) according to the manufacturer’s protocol.
MicroarrayArraystar Human LncRNA Microarray V3.0 is designed for the global profiling of human lncRNAs and protein-coding transcripts,which is updated from the previous Microarray V2.0. About 30,586 lncRNAs and 26,109 coding transcripts can be detected by V3.0 microarray. Sample labeling and array hybridization were performed according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology) with minor modifications. Briefly,mRNA was purified from total RNA after removal of rRNA (mRNA-ONLYTM Eukaryotic mRNA Isolation Kit,Epicentre). Then,each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3’ bias utilizing a random priming method. The labeled cRNAs were purified by RNeasy Mini Kit (Qiagen). The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured by NanoDrop ND-1000. One microgram of each labeled cRNA was fragmented by adding 5 μL of 10 × blocking agent and 1 μL of 25 × fragmentation buffers,and then the mixture was heated at 60 °C for 30 min and 25 μL of 2 × GE hybridization buffer was added to dilute the labeled cRNA. Then,50 μL of hybridization solution was dispensed into the gasket slide and assembled onto the lncRNA expression microarray slide. The slides were incubated for 17 h at 65 °C in an Agilent Hybridization Oven. The hybridized arrays were washed,fixed,and scanned using the Agilent DNA Microarray Scanner (part number G2505C).
Real-time Quantitative Reverse-transcription PCR (qRT-PCR )About 2 μg of total RNA was reverse-transcribed into first-strand cDNA pool using SuperScriptTM reverse transcriptase and random primers (ABI). Q-PCR was performed using the SYBR Green I Q-PCR kit (Life Sciences Solutions Group,Thermo Fisher Scientific) on a Bio-Rad CFX system. Specific primers are shown in Table S1 (in the website of BES,www.besjournal.com).
Construction of the Coding-noncoding Gene Co-expression NetworkThe network construction procedures include (i) preprocessing data: the same coding gene with different transcripts takes the median value representing the gene expression values,without special treatment of lncRNA expression value; (ii) screening data: removing the subset of data according to the lists that show the differential expression of lncRNAs and mRNAs; (iii) calculation of Pearson correlation coefficient (PCC) and using R value to calculate PCC between lncRNAs and coding genes; and (iv) screening by PCC,selecting the part that has PCC ≥0.99 as significant,and drawing the coding-noncoding gene co-expression network by Cytoscape.
Prediction of Coding PotentialThe coding potential of lncRNAs was evaluated by Coding Potential Calculator (CPC) with higher accuracy and calculation speed[19]. The web-based interface of CPC is at http://cpc.cbi.pku.edu.cn,the examined sequences can be pasted directly into the input box,the CPC server runs and results will be shown in the browser once the computation is finished,and the output score is termed as support vector machine (SVM) score. The transcripts with scores between −1 and 1 are marked as ‘weak noncoding’ and ‘weak coding’ respectively. The farther away the score is from zero,the more reliable the prediction is.
Plasmids,Cell Culture,and TransfectionpcDNA3.1-ARG was constructed through restric- tion digestion and ligation-mediated cloning and was verified by restriction digestion and sequencing prior to use. The PCR primers were AAGCTTCTGTGTGCACT (HindIII) and GAATTCTTACTGTAGTAA (EcoRI). HeLa cells were purchased from China infrastructure of cell line resources and grown at 37 °C,with 5% CO2,in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% (v/v) penicillin-streptomycin. Cell transfection was performed according to the manufacturer’s protocol (Vigorous).
Measurement of Intracellular Superoxide LevelsSuperoxide production was detected using the fluorescent 2’,7’-dichlorofluorescein (DCF) obtained from Vigorous according to a previous study[20]. HeLa cells were cultured in six-well plates,transfected with the indicated plasmids,and 48 h later washed with PBS and labeled with 10 μmol/L DCF in the culture plates at 37 °C for 15 min in PBS. The culture plates were placed on ice to stop the labeling,trypsinized,and resuspended in ice-cold PBS. Samples were analyzed using a flow cytometer. Data analysis was performed using c-flow software,and the mean fluorescence intensity was used to quantify the responses. A minimum of 10,000 cells were acquired for each sample.
Data AnalysisAgilent Feature Extraction software (version 11.0.1.1) was used to analyze the acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX v12.0 software package (Agilent Technologies). After quantile normalization of the raw data,lncRNAs and mRNAs with at least three out of six samples having flags in Present or Marginal (‘All Targets Value’) were chosen for further data analysis. Differentially expressed lncRNAs and mRNAs with statistical significance between the two groups were identified through volcano plot filtering. Hierarchical clustering was performed using the Agilent GeneSpring GX software (version 12.0). GO analysis and pathway analysis were performed in the standard enrichment computational method. All data were expressed as mean±SD. P value of <0.05 was considered as statistically significant.
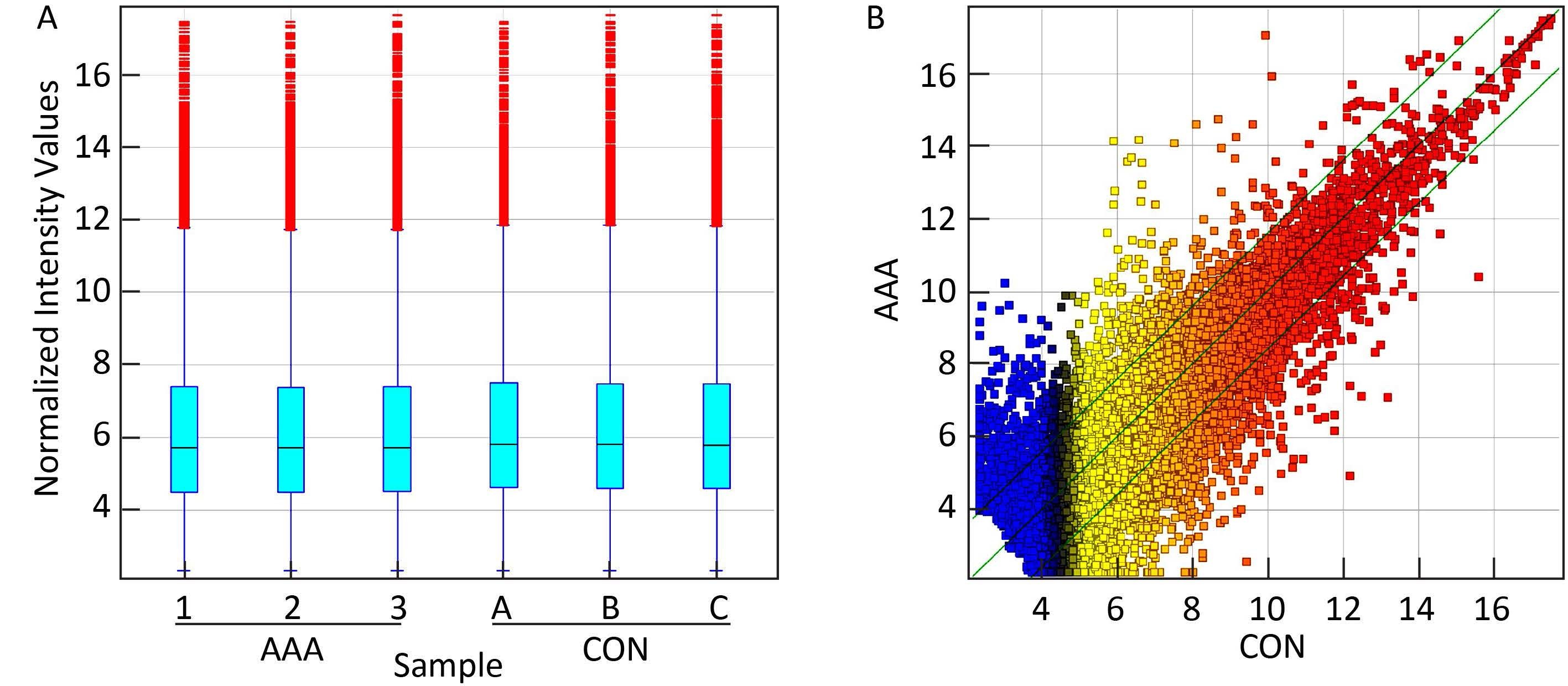
RESULTS LncRNA Expression Profiles in AAATo analyze the potential biological functions of lncRNAs in AAA,we determined the lncRNA expression profile in human AAA aortic tissues through microarray technology (Figure 1). From the lncRNA expression profile,tens of thousands of lncRNAs (26,946) collected from human lncRNA databases,including RefSeq_NR,UCSC_knowngene,Ensembl,H-invDB,Fantom,Fantom_stringent,NRED,RNAdb,misc_lncRNA,UCR,and lncRNA,could be examined in normal and AAA aortic tissues (Table S2 in the website of BES,www.besjournal.com). Furthermore,we identified 3,688 lncRNAs that were significantly differentially expressed between the three AAA patients and three normal subjects (≥3-fold,P<0.05,Table S3 in the website of BES,www.besjournal.com). Among these candidates,1,582 lncRNAs were upregulated more than 3-fold in the AAA group compared to that in the normal aortic tissue group,while 2,106 lncRNAs were downregulated more than 3-fold (P<0.05,Table S3 in the website of BES,www.besjournal.com). ENST00000566575 (log2-fold change T/N=306.7137) was the most significantly downregulated lncRNA,while ENST00000512263 (log2-fold change T/No= 148.8989) was the most significantly upregulated lncRNA (Table 1).
|
Download:
|
| Figure 1 Comparison of lncRNA expression profile between the AAA aortic samples and the normal aortic samples. (A) The box plot is a convenient method to quickly compare the distribution of lncRNAs. After normalization,the distributions of log 2 ratios among the tested samples are almost similar. (B) The scatterplot is a visualization method that is useful for assessing the variation (or reproducibility) between the AAA and normal aortic tissues compared by microarrays. The values of X and Y axes in the scatterplot are averaged normalized values in each group (log 2 scaled). The green lines are fold change lines (the default fold change value given is 3.0). The lncRNAs above the top green line and below the bottom green line indicate more than 3-fold change of lncRNAs between pairs. | |
|
|
Table 1 A Collection of Differential lncRNAs Detected by Microarray in AAA and Control Tissues |
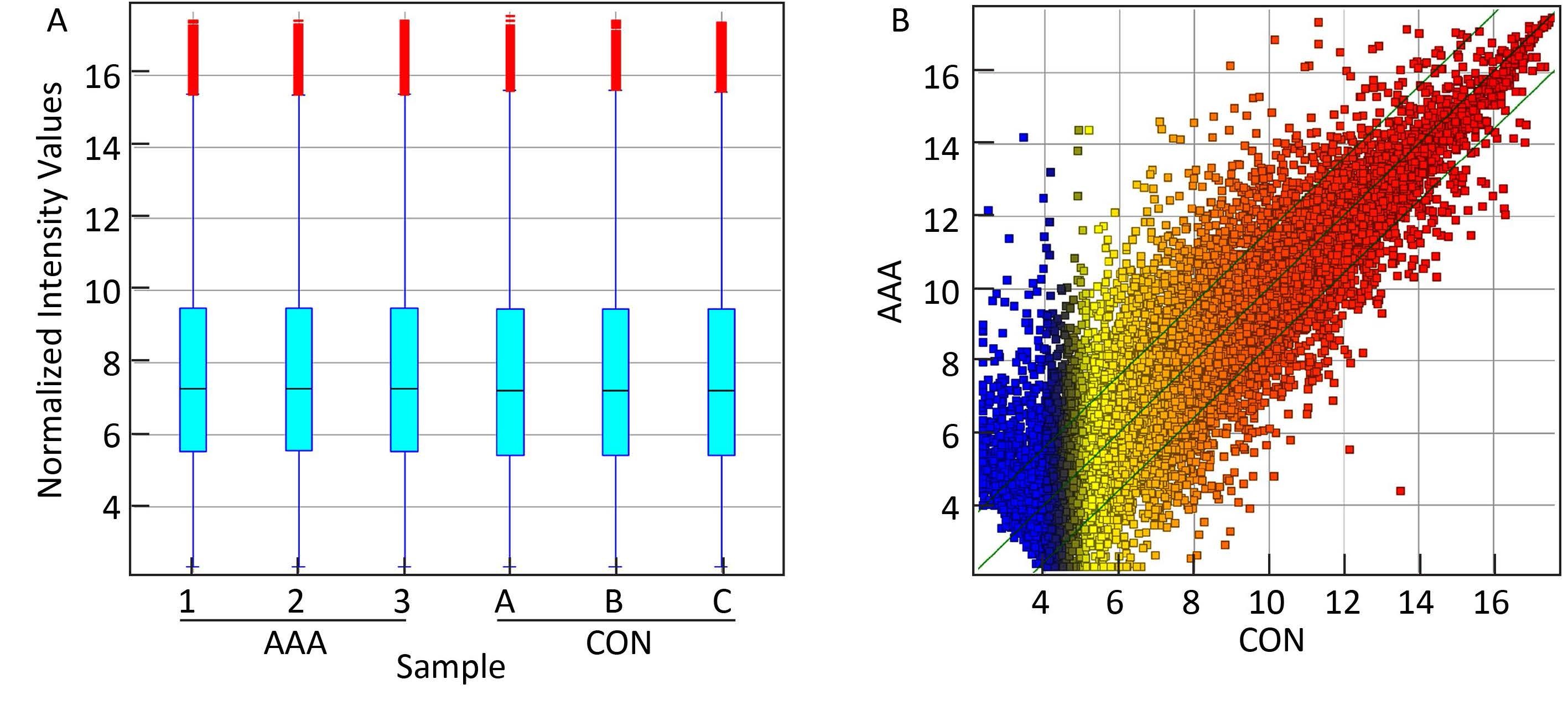
The mRNA expression profile in human AAA tissues was also examined through microarray analysis (Figure 2). Up to 22,266 coding transcripts could be detected in the three AAA and three normal aortic tissue samples through 30,215 coding transcripts probes (Table S4 in the website of BES,www.besjournal.com). Among the three pairs of samples,1,833 mRNAs were upregulated in AAA patients compared to those in the matched normal tissues,while 1,174 mRNAs were downregulated (Table S5 in the website of BES,www.besjournal.com).
|
Download:
|
| Figure 2 Comparison of mRNA expression profile between the AAA aortic samples and normal aortic samples. (A) The box plot is a convenient method to quickly compare the distribution of mRNAs. After normalization,the distributions of log2 ratios among the tested samples are almost similar. (B) The scatterplot is a visualization method that is useful for assessing the variation (or reproducibility) between the AAA and normal aortic tissues compared by microarrays. The values of X and Y axes in the scatterplot are averaged normalized values in each group (log 2 scaled). The green lines are fold change lines (the default fold change value given is 3.0). The lncRNAs above the top green line and below the bottom green line indicate more than 3-fold change of lncRNAs between pairs. | |
GO and pathway analyses showed that the differentially expressed mRNAs might be involved in different signaling pathways of biological processes,cellular components,and molecular functions. The upregulated mRNAs primarily correlated with the immune system processes and immune response in biological processes and with cytokine and chemokine activity involved in molecular functions (Table 2). We also found that some upregulated differentially expressed mRNAs correlated with the extracellular space and region in the cellular components (Table 2),and the downregulated mRNAs were primarily associated with muscle system process and contraction,muscle structure and tissue development in biological processes,contractile fibers and myofibrils in the cellular components,and cytoskeletal protein binding involved in molecular functions (Table 2). These signaling pathways may be involved in the pathogenesis of AAA. In addition,our results showed that uncoupling protein 1 (UCP-1) was the most significantly downregulated molecule in AAA and MMPs,including MMP12,MMP13,MMP7,and MMP3,were obviously upregulated in AAA (Table 3).
|
|
Table 2 Functional Classification of Target Genes by GO |
|
|
Table 3 A Collection of Differential mRNAs Detected by Microarray in AAA and Control Tissues |
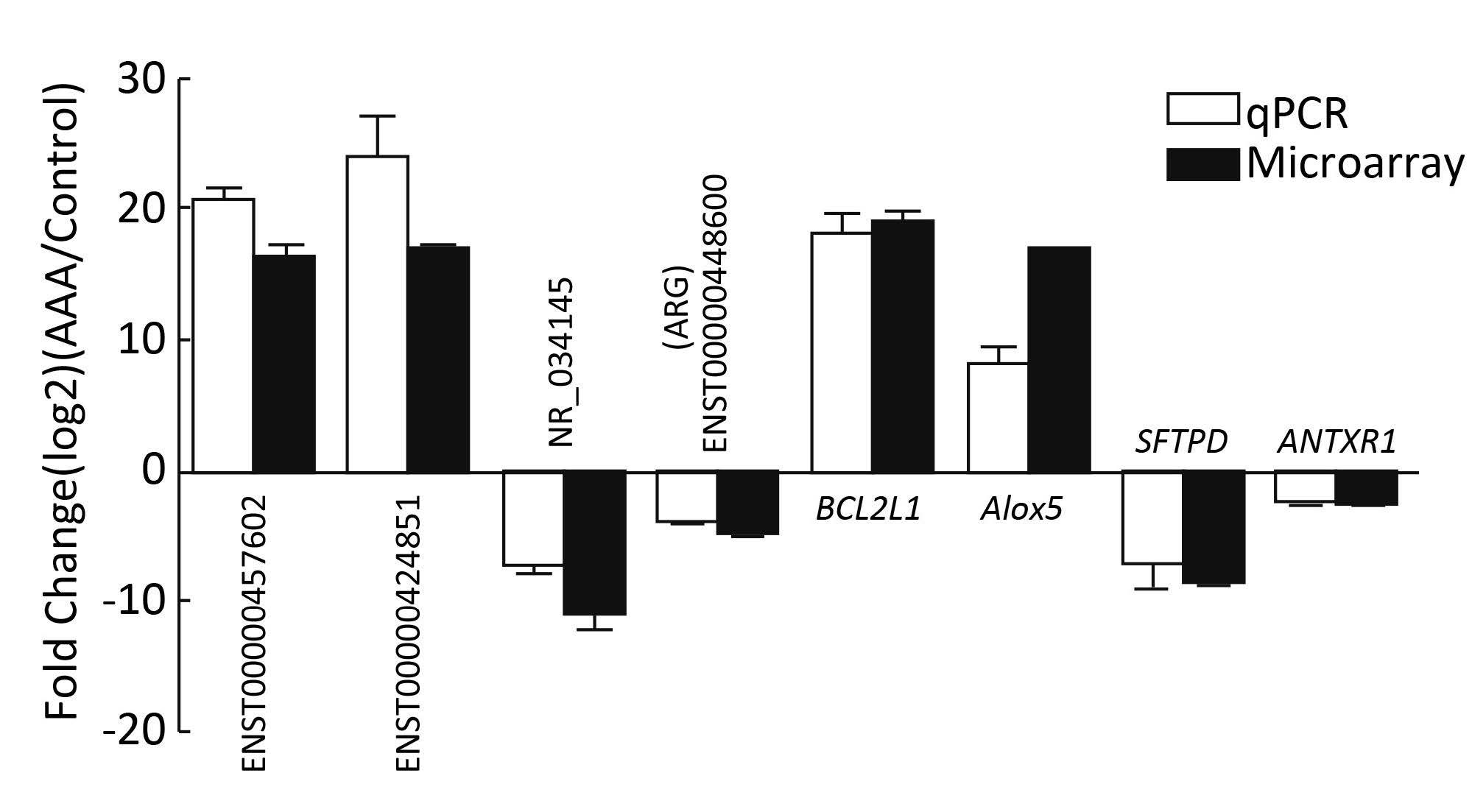
To further determine the accuracy of the microarray results,we examined the expression of some lncRNAs and mRNAs in AAA and control tissues using qRT-PCR. Compared with normal tissue samples,the lncRNAs of ENST00000457602 and ENST00000424851 were upregulated and the lncRNAs of NR_034145 and ENST00000448600 were downregulated,while the mRNAs of BCL-2-like protein 1 (BCL2L11) and ALOX5 were upregulated,and the mRNAs of anthrax toxin receptor 1 isoform 1 (ANTXR1) and pulmonary surfactant-associated protein D (SFTPD) were downregulated in AAA tissue samples (Figure 3),which are consistent with the microarray results. These data support a strong consistency between the qRT-PCR result and microarray data.
|
Download:
|
| Figure 3 Comparison between microarray data and qRT-PCR results. Total RNA from AAA (n=6) and control tissues (n=6) was extracted and analyzed by qRT-PCR for each sample. | |
Although the vast majority of lncRNAs described in the literature remain to be studied in a mechanistic detail,previous studies have indicated that lncRNAs participate in biological processes by regulating the expression of their target genes[13]. Long intergenic noncoding RNAs (lincRNAs) have been extensively studied for their expression feature and conservation among species[21]. In the current study,10,608 differentially expressed lincRNAs could be found in normal and AAA aortic tissues (Table S6 in the website of BES,www.besjournal.com). Furthermore,we identified 1,284 differentially expre- ssed lincRNAs and nearby coding gene pairs (distance <300 kb,P<0.05,shown in Table S7 in the website of BES,www.besjournal.com). A previous study indicated that a majority of lncRNAs with enhancer-like functions are found in human cell lines and depletion of these lncRNAs led to decreased expression of their neighboring protein-coding genes[22]. Deduced from bioinformatics analysis,we predicted 1,611 lncRNA candidates with enhancer-like functions in normal and AAA aortic tissues (shown in Table S8 in the website of BES,www.besjournal.com) and identified 206 differentially expressed enhancer-like lncRNAs and their potential target coding gene pairs (distance <300 kb,P<0.05,shown in Table S9 in the website of BES,www.besjournal.com).
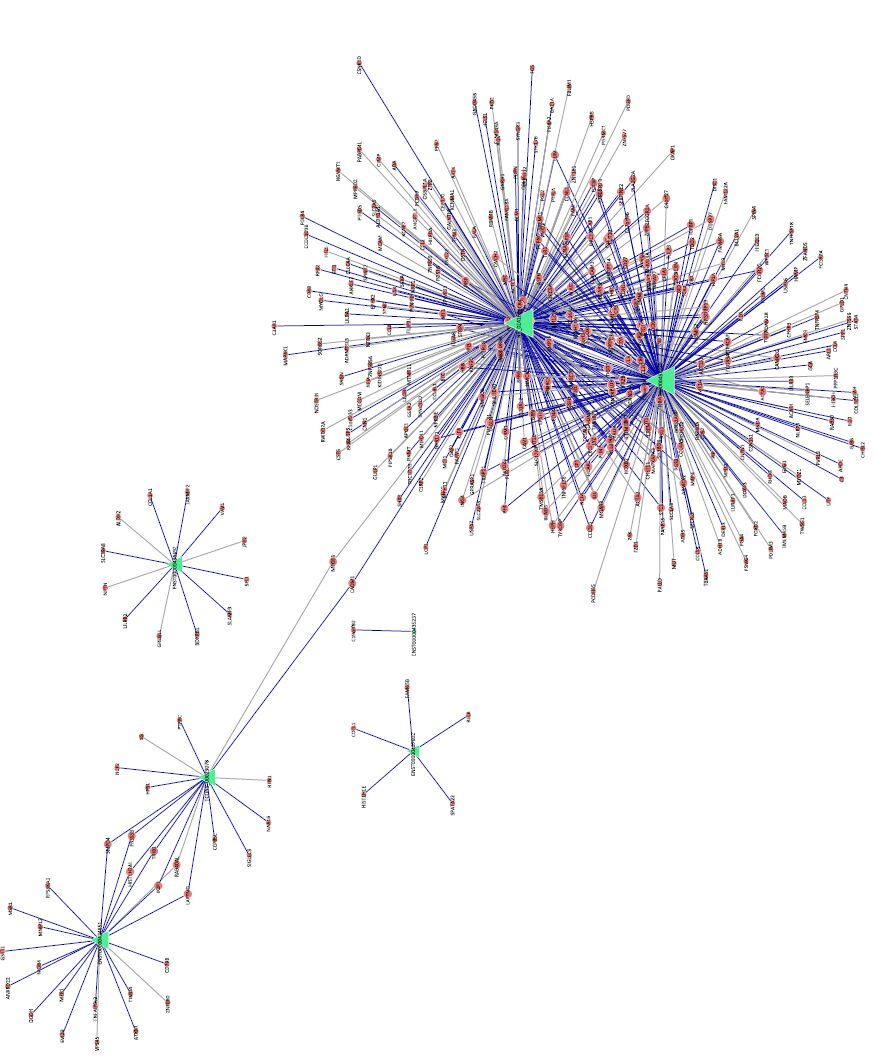
A coding-noncoding gene co-expression network (CNC network) was constructed based on the correlation analysis between the differentially expressed lncRNAs and mRNAs. Seven obviously differentially expressed lncRNAs were selected to draw the network using Cytoscape,based on the PCCs of lncRNAs and mRNAs not less than 0.[99] A total of 416 network nodes made associated 573 network pairs of co-expression of lncRNAs and mRNAs,and 371 pairs showed a positive correlation and 202 pairs showed a negative correlation. Among the CNC network,the lncRNAs of NR_024160 and ENST00000457239 correlated with 285 and 231 mRNAs,respectively (Figure 4 and Table S10 in the website of BES,www.besjournal.com). ALOX5,an enzyme that catalyzes the first committed step in the metabolic pathway leading to the synthesis of proinflammatory LTs[23],correlated with the lncRNAs of NR_024160 and ENST00000457239 (Table S10 in the website of BES,www.besjournal.com).
|
Download:
|
| Figure 4 Construction of the coding-noncoding gene co-expression network. The red circular node represents mRNA and the green triangle node represents lncRNA. The blue lines indicate a positive correlation and the gray line indicates a negative correlation. | |
Since the possible functions of lncRNAs can often be inferred based on existing examples and their relative genomic organization[24],we found that ENST00000448600 (termed as ALOX5 regulatory gene,lnc-ARG) is located within the upstream of ALOX5 and preliminarily investigated the role of lnc-ARG in regulating ALOX5. We analyzed the protein-coding potential of lnc-ARG using computational analysis[19]. The indicative SVM score of lnc-ARG was −0.918424,which is sorted as a noncoding transcript. Furthermore,our results showed that lnc-ARG overexpression slightly decreased the mRNA level of ALOX5 and ROS production in HeLa cells (Figure 5A and 5B).

|
Download:
|
| Figure 5 Lnc-ARG overexpression reduced ALOX5 expression and ROS production. (A) The mRNA level of ALOX5 was inhibited by lnc-ARG overexpression. (B) Lnc-ARG overexpression reduced ROS production. P<0.05,**P<0.01,***P<0.001. | |
Exploratory studies have so far identified a few lncRNAs associated with cardiovascular diseases,including metastasis-associated lung adenocarcinoma transcript 1 (MALAT1),lincRNA predicting cardiac remodeling (LIPCAR),cardiac apoptosis-related lncRNA (CARL),and so on[25]. However,specific studies analyzing the lncRNAs in aortic aneurysm are still lacking[25-26]. In this study,we first describe the expression profiles of human lncRNAs in AAA by microarray and identify a set of differentially expressed lncRNAs in AAA in comparison with NT,which provides candidates for further insight into the pathogenesis of AAA.
Compared to normal tissues,microarray techniques describe a set of differentially expressed lncRNAs,with 1,582 upregulated and 2,106 downregulated lncRNAs in AAA tissues. Regarding the mRNA expression profile in human AAA,we also found 1,833 upregulated mRNAs and 1,174 downregulated mRNAs in AAA tissues compared with the matched normal tissues by microarray analysis. The aim of identifying the putative functions of nearby genes of lncRNAs is to explore the functional roles of lncRNAs. A large number of lincRNAs and lncRNAs with enhancer-like functions have been explored,the functions of which are related to cis or trans transcriptional regulation,translational control,splicing regulation,and other post-transcriptional regulation[27-30]. In the current study,according to the genomic location and functional prediction,we identified 1,284 differentially expressed lincRNAs and nearby coding gene pairs and 206 differentially expressed enhancer-like lncRNAs located within the regulatory regions of their neighbor protein-coding genes.
Functional enrichment analysis showed that the upregulated differentially expressed mRNAs primarily correlated with the immune system processes and immune response in biological processes and with cytokine and chemokine activity involved in molecular functions. Studies on human AAA tissues have shown extensive inflammatory infiltrates containing macrophages,lymphocytes,and mast cells in both the media and adventitia,and increasing aneurysm diameter was associated with a higher density of inflammatory cells in the adventitia[8, 31]. Previous studies indicate that UCPs play a major role in thermoregulation,heat generation,and maintenance of basal metabolic rate and participate in the antioxidant protection of the body[32]. Moreover,energy metabolism and oxidation stress are related to the pathology of AAA. In the present study,our results showed that UCP-1 is the most significantly downregulated molecule in AAA and indicated that UCP-1 may have important functions in the pathogenesis of AAA.
Increasing evidence has indicated that lncRNAs have important biological functions such as controlling the expression of protein-coding genes and some lncRNAs could act as cofactors to modulate transcription factor activity to induce expression of adjacent protein-coding genes[33],while some lncRNAs could interact with the initiation complex to repress the expression of downstream genes[28-29]. Collective studies indicate that lncRNAs are involved in human diseases such as tumors,Alzheimer’s disease,and cardiovascular diseases[25, 34-36]. Recently,Huang et al. in 2016 demonstrated that the expression of several lncRNAs is dynamically regulated in ischemic cardiomyopathy. Furthermore,lncRNAs regulated the expression and function of the extracellular matrix and cardiac fibrosis during the development of ischemic cardiomyopathy[37]. In addition,a previous study showed that HIF1A-AS1 mediates the pro-apoptosis and antiproliferative responses induced by the Brahma-related gene 1 (BRG1) in VSMCs,indicating that the lncRNA HIF1A-AS1 plays an important role in the pathophysiology of VSMCs[38]. Furthermore,the expression of several lncRNAs was found to be regulated by Ang II in VSMCs and the lncRNA AngII362 mediated the proliferation of VSMCs by acting on miR221/222[39]. There are some candidate lncRNAs that may play a role in inflammation and extracellular matrix remodeling and fibrosis[14]. In this study,bioinformatics analysis was performed to predict the targeted mRNAs of lncRNAs by combining differentially expressed mRNAs and lncRNAs and to construct the co-expression networks of lncRNAs and mRNAs in AAA. We found that lnc-ARG overexpression slightly inhibited the mRNA level of its downstream gene,ALOX5. ALOX5 was found to be present in human atherosclerotic aorta and the coronary and carotid arteries and may play a significant role in modifying the pathogenesis of atherosclerosis[40]. Atherosclerosis represents an important chronic inflammatory process associated with several pathophysiological reactions in the vascular wall[41]. LTs can induce proinflammatory signaling and plays an important role in atherosclerosis through the activation of specific BLT and CysLT receptors,further causing vascular wall morphological alterations such as early lipid retention and accumulation of foam cells,development of intimal hyperplasia,and advancing atherosclerotic lesions,eventually,resulting in the rupture of atherosclerotic plaque[40]. A previous study showed that ALOX5 plays a critical role in 4-hydroxynonenal-induced ROS generation in murine macrophages through the activation of NADPH oxidase[42]. In the present study also,lnc-ARG overexpression slightly reduced ROS production.
In summary,the differentially expressed lncRNAs identified in our study may play important roles in the pathology and formation of AAA. Analyzing the co-expression networks of lncRNAs and mRNAs in AAA will help provide further insight into the pathogenesis of AAA. Because of the difficulty in obtaining tissue samples,the small sample size is a limitation of this study and a further study with large sample size is planned to verify the relevant results and to clarify the detailed molecular mechanism of lncRNA-mediated regulation of their potential coding genes in AAA.
ACKNOWLEDGMENTS
We thank all the donors who participated in this program. We also thank all those who devoted to the Microarray service at KangChen Bio-technology Company at Shanghai.
| 1. | Lederle FA, Johnson GR, Wilson SE, et al. The aneurysm detection and management study screening program:validation cohort and final results. Aneurysm Detection and Management Veterans Affairs Cooperative Study Investigators[J]. Arch Intern Med , 2000, 160 :1425–30. doi:10.1001/archinte.160.10.1425 |
| 2. | Miyake T, Morishita R. Pharmacological treatment of abdominal aortic aneurysm[J]. Cardiovasc Res , 2009, 83 :436–63. doi:10.1093/cvr/cvp155 |
| 3. | Morris DR, Biros E, Cronin O, et al. The association of genetic variants of matrix metalloproteinases with abdominal aortic aneurysm:a systematic review and meta-analysis[J]. Heart , 2014, 100 :295–302. doi:10.1136/heartjnl-2013-304129 |
| 4. | Tamarina NA, McMillan WD, Shively VP, et al. Expression of matrix metalloproteinases and their inhibitors in aneurysms and normal aorta[J]. Surgery , 1997, 122 :271–62. |
| 5. | Defawe OD, Colige A, Lambert CA, et al. TIMP-2 and PAI-1 mRNA levels are lower in aneurysmal as compared to athero-occlusive abdominal aortas[J]. Cardiovasc Res , 2003, 60 :205–13. doi:10.1016/S0008-6363(03)00513-3 |
| 6. | Abisi S, Burnand KG, Waltham M, et al. Cysteine protease activity in the wall of abdominal aortic aneurysms[J]. J Vasc Surg , 2007, 46 :1260–6. doi:10.1016/j.jvs.2007.08.015 |
| 7. | Lindholt JS, Erlandsen EJ, Henneberg EW. Cystatin C deficiency is associated with the progression of small abdominal aortic aneurysms[J]. Br J Surg , 2001, 88 :1472–5. doi:10.1046/j.0007-1323.2001.01911.x |
| 8. | Freestone T, Turner RJ, Coady A, et al. Inflammation and matrix metalloproteinases in the enlarging abdominal aortic aneurysm[J]. Arterioscler Thromb Vasc Biol , 1995, 15 :1145–51. doi:10.1161/01.ATV.15.8.1145 |
| 9. | Miller FJ, J r., Sharp WJ, Fang X. Oxidative stress in human abdominal aortic aneurysms:a potential mediator of aneurysmal remodelin[J]. Arterioscler Thromb Vasc Biol , 2002, 22 :560–5. doi:10.1161/01.ATV.0000013778.72404.30 |
| 10. | McCormick ML, Gavrila D, Weintraub NL. Role of oxidative stress in the pathogenesis of abdominal aortic aneurysms[J]. Arterioscler Thromb Vasc Biol , 2007, 27 :461–9. doi:10.1161/01.ATV.0000257552.94483.14 |
| 11. | Zhang J, Schmidt J, Ryschich E, et al. Inducible nitric oxide synthase is present in human abdominal aortic aneurysm and promotes oxidative vascular injury[J]. J Vasc Surg , 2003, 38 :360–7. doi:10.1016/S0741-5214(03)00148-4 |
| 12. | Ng SY, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system[J]. Trends Genet , 2013, 29 :461–8. doi:10.1016/j.tig.2013.03.002 |
| 13. | Kung JT, Colognori D, Lee JT. Long noncoding RNAs:past, present, and future[J]. Genetics , 2013, 193 :651–69. doi:10.1534/genetics.112.146704 |
| 14. | Peters T, Schroen B. Missing links in cardiology:long non-coding RNAs enter the arena[J]. Pflugers Arch , 2014, 466 :1177–87. doi:10.1007/s00424-014-1479-1 |
| 15. | Maegdefessel L, Azuma J, Toh R, et al. MicroRNA-21 blocks abdominal aortic aneurysm development and nicotine-augmented expansion[J]. Sci Transl Med , 2012, 4 :122ra122. |
| 16. | Leeper NJ, Raiesdana A, Kojima Y, et al. MicroRNA-26a is a novel regulator of vascular smooth muscle cell function[J]. J Cell Physiol , 2011, 226 :1035–43. doi:10.1002/jcp.22422 |
| 17. | Maegdefessel L, Azuma J, Toh R, et al. Inhibition of microRNA-29b reduces murine abdominal aortic aneurysm development[J]. J Clin Invest , 2012, 122 :497–506. doi:10.1172/JCI61598 |
| 18. | Kin K, Miyagawa S, Fukushima S, et al. Tissue- and plasma-specific MicroRNA signatures for atherosclerotic abdominal aortic aneurysm[J]. J Am Heart Assoc , 2012, 1 :e000745. |
| 19. | Kong L, Zhang Y, Ye ZQ, et al. CPC:assess the protein-coding potential of transcripts using sequence features and support vector machine[J]. Nucleic Acids Res , 2007, 35 :W345–9. doi:10.1093/nar/gkm391 |
| 20. | Eruslanov E, Kusmartsev S. Identification of ROS using oxidized DCFDA and flow-cytometry[J]. Methods Mol Biol , 2010, 594 :57–72. doi:10.1007/978-1-60761-411-1 |
| 21. | Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs[J]. RNA Biol , 2013, 10 :925–33. |
| 22. | Orom UA, Derrien T, Beringer M, et al. Long noncoding RNAs with enhancer-like function in human cells[J]. Cell , 2010, 143 :46–58. doi:10.1016/j.cell.2010.09.001 |
| 23. | Anwar Y, Sabir JS, Qureshi MI, et al. 5-lipoxygenase:a promising drug target against inflammatory diseases- biochemical and pharmacological regulation[J]. Curr Drug Targets , 2014, 15 :410–22. doi:10.2174/1389450114666131209110745 |
| 24. | Knauss JL, Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function[J]. Neuroscience , 2013, 235 :200–14. doi:10.1016/j.neuroscience.2013.01.022 |
| 25. | Duggirala A, Delogu F, Angelini TG. Non coding RNAs in aortic aneurysmal disease[J]. Front Gene , 2015, 6 :125. |
| 26. | Jiang X, Ning Q. The emerging roles of long noncoding RNAs in common cardiovascular diseases[J]. Hypertens Res , 2015, 38 :375–9. doi:10.1038/hr.2015.26 |
| 27. | Willingham AT, Orth AP, Batalov S, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT[J]. Science , 2005, 309 :1570–3. doi:10.1126/science.1115901 |
| 28. | Martianov I, Ramadass A, Serra Barros A, et al. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript[J]. Nature , 2007, 445 :666–70. doi:10.1038/nature05519 |
| 29. | Wang X, Arai S, Song X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription[J]. Nature , 2008, 454 :126–30. doi:10.1038/nature06992 |
| 30. | Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs:insights into functions[J]. Nat Rev Genet , 2009, 10 :155–9. doi:10.1038/nrg2521 |
| 31. | Tsuruda T, Kato J, Hatakeyama K, et al. Adventitial mast cells contribute to pathogenesis in the progression of abdominal aortic aneurysm[J]. Circ Res , 2008, 102 :1368–77. doi:10.1161/CIRCRESAHA.108.173682 |
| 32. | Mailloux RJ, Harper ME. Uncoupling proteins and the control of mitochondrial reactive oxygen species production[J]. Free Radic Biol Med , 2011, 51 :1106–15. doi:10.1016/j.freeradbiomed.2011.06.022 |
| 33. | Feng J, Bi C, Clark BS, et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator[J]. Genes Dev , 2006, 20 :1470–84. doi:10.1101/gad.1416106 |
| 34. | Shi X, Sun M, Liu H. Long non-coding RNAs:a new frontier in the study of human diseases[J]. Cancer Lett , 2013, 339 :159–66. doi:10.1016/j.canlet.2013.06.013 |
| 35. | Esteller M. Non-coding RNAs in human disease[J]. Nat Rev Genet , 2011, 12 :861–74. |
| 36. | Tang JY, Lee JC, Chang YT, et al. Long noncoding RNAs-related diseases, cancers, and drugs[J]. TheScientificWorldJournal , 2013, 2013 :943539. |
| 37. | Huang ZP, Ding Y, Chen J, et al. Long non-coding RNAs link extracellular matrix gene expression to ischemic cardiomyopathy. Cardiovasc Res, 2016. |
| 38. | Wang S, Zhang X, Yuan Y, et al. BRG1 expression is increased in thoracic aortic aneurysms and regulates proliferation and apoptosis of vascular smooth muscle cells through the long non-coding RNA HIF1A-AS1 in vitro[J]. Eur J Cardiothorac Surg , 2015, 47 :439–46. doi:10.1093/ejcts/ezu215 |
| 39. | Leung A, Trac C, Jin W, et al. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells[J]. Circ Res , 2013, 113 :266–78. doi:10.1161/CIRCRESAHA.112.300849 |
| 40. | Riccioni G, Zanasi A, Vitulano N, et al. Leukotrienes in atherosclerosis:new target insights and future therapy perspectives[J]. Mediators Inflamm , 2009, 2009 :737282. |
| 41. | Ross R. Atherosclerosis——an inflammatory disease[J]. N Engl J Med , 1999, 340 :115–26. doi:10.1056/NEJM199901143400207 |
| 42. | Yun MR, Park HM, Seo KW, et al. 5-Lipoxygenase plays an essential role in 4-HNE-enhanced ROS production in murine macrophages via activation of NADPH oxidase[J]. Free Radic Res , 2010, 44 :742–50. doi:10.3109/10715761003758122 |
 2016, Vol. 29
2016, Vol. 29


