叶绿素是植物体内重要的光合色素,以色素蛋白复合体形式存在于植物类囊体中,在光合作用中起捕获并传递光能的作用。植物叶绿素主要由叶绿素a(chlorophyll a,Chl a)和叶绿素b(chlorophyll b,Chl b)组成,两者结构类似,包含卟啉环、卟啉环中央的镁原子和长脂肪烃侧链,它们仅在吡咯环Ⅱ的附加基团有差异: 前者是甲基,后者是醛基[1]。叶绿素吸收光能,通过光合作用为植物体发育提供能量,但过量的叶绿体存在会产生大量自由基,加速植物细胞死亡[2]。叶绿素降解产物具有抗氧化的作用,可以维持或缓解细胞活性。因此,叶绿素的适时降解具有重要的生物学意义。
植物叶绿素在体内以动态形式存在。据统计,全球每年约有109 t叶绿素在植物中被降解[3]。其中,叶绿素降解速率的变化是植物发育的重要调节剂和指示剂。如年龄增加、细胞死亡、外界温度的剧烈变化、虫害及病原体等都会加速叶绿素降解[4, 5]。丰富的叶绿素结合蛋白在降解过程中分解形成氮素被重新利用,这部分再利用的氮素对植物的生长发育及生存繁衍意义重大[6]。此外,衰老阶段出现的叶绿素降解缓慢或者不降解的现象,即“滞绿”现象,是作物高产的重要指标之一[7]。因此,阐明植物叶绿素降解代谢及调控机理对明确植物的生长发育调控、作物的品种改良和农业生产控制具有重要的理论和实践意义。自20世纪90年代对叶绿素降解产物结构的解析和相关酶基因的克隆,叶绿素降解过程逐渐明晰[6]。近年,随着结构生物学、基因组测序和生物信息学的发展,对植物叶绿素降解途径的分子调控机制研究已经取得了巨大进展。本文主要对近年来叶绿素降解代谢和调控机理方面的研究进展进行综述,并对本领域未来研究方向进行了展望,以期为农业生产和作物育种提供理论依据。
1 叶绿素的降解途径1991年大麦中非荧光叶绿素代谢物(nonfluore-scent chl catabolites,NCCs)的发现,加速了对叶绿素代谢研究[8]。到目前为止,植物中已发现15种结构的NCCs[9]。进一步对底物脱镁叶绿酸a(phei-de a)分析发现红色叶绿素代谢物(red chlorophyll catabolites,RCCs)和初级荧光叶绿素代谢物(prim-ary fluorescent chlorophyll catabolite,pFCC)[10]。
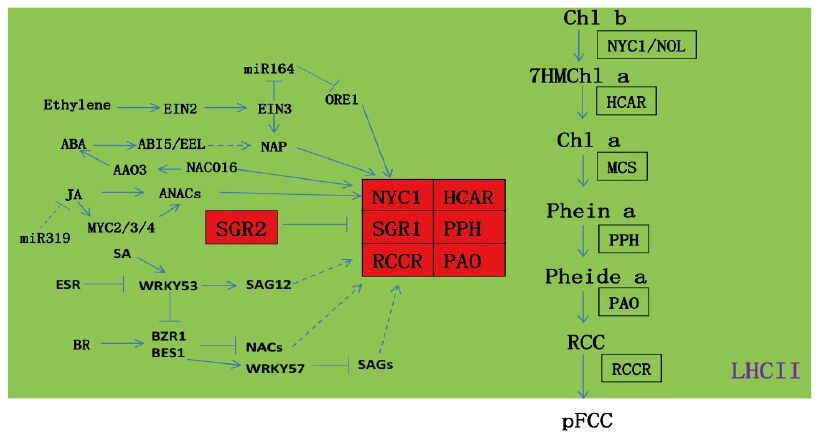
植物叶绿素a占总叶绿素的比重较大,在降解的初始阶段,叶绿素a被叶绿素酶(chrolophyllase,CS)催化形成脱植基叶绿素a(chlide a)和叶绿醇;脱植基叶绿素a后经脱镁螯合酶(Mg-dechelatase,MDCase)脱去镁离子形成脱镁叶绿素a(pheophytin a),并保持卟啉大环结构[6]。脱植基叶绿素a可在叶绿素加氧酶的作用下合成叶绿素b,叶绿素b在叶绿素b还原酶的作用下又可还原成叶绿素a,这种转换称为“叶绿素循环”。在脱镁叶酸酶加氧酶和红色叶绿素代谢产物还原酶的作用下,脱植基叶绿素a降解形成pFCC并运输到液泡中,期间会形成一种不稳定代谢产物红色叶绿素代谢产物(RCC),由于卟啉环被打开,绿色随之消失,最后pFCC在液泡中部分高度修饰形成荧光叶绿素代谢物hFCCs(hypermodifided FCCs),或修饰形成mFCCs(modified FCCs)并异构成NCCs;至此,叶绿素从绿色被降解成无色化合物,完成叶绿素的降解。此叶绿素降解途径,称之为脱镁叶绿素a加氧酶(pheide a oxygenase,PAO)途径[10](图 1),在植物中高度保守,其活性只出现在叶片衰老和果实成熟的阶段。
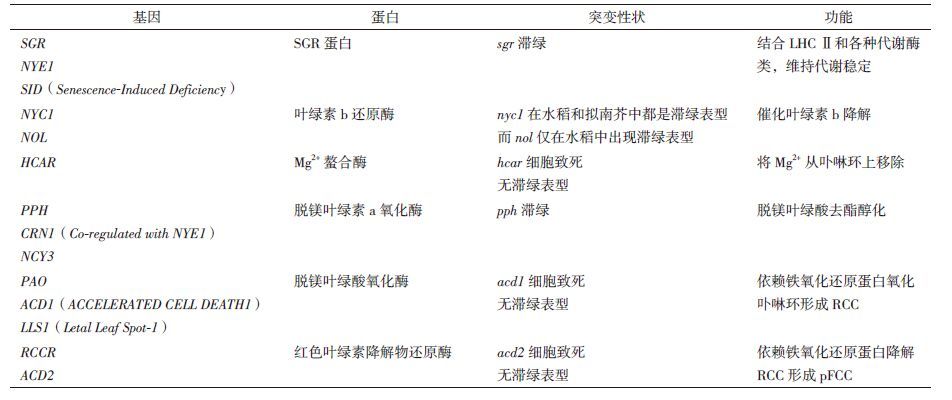
2 叶绿素降解主要酶类及基因调控 2.1 叶绿素降解主要代谢酶类及基因叶绿素降解受体内酶类代谢调控。在拟南芥和水稻中已鉴定了6种叶绿素代谢酶(Chl catabolic enzymes,CCEs),包括NON-YELLOW COLORING1(NYC1)和NYC1-LIKE(NOL)[11]编码的叶绿素b还原酶、7-羟甲基叶绿素还原酶(7-Hydroxymethyl chlorophyll a reductase,HCAR)[12]、脱镁叶绿素酶(pheophytinase,PPH)[13]、PAO[14]和红色代谢物还原酶(red chlorophyll catabolite reductase,RCCR)[15]。除上述主要酶类代谢基因外,拟南芥PAO途径中,STAY-GREEN(SGR)和METHYLESTERASE FAMILY MEMBER 16(MES16)也参与调控叶绿素降解[13, 16-19]。
水稻中,NYC1和NOL基因编码叶绿素b还原酶,两者共同定位在内囊体膜上,形成叶绿素b还原酶复合体,催化叶绿素b的降解[11]。水稻滞绿突变体nyc1在叶片衰老过程中,叶绿素和捕光色素复合体(light-harvesting pigment protein complex II,LHCII)中类胡萝卜素的降解同时受到抑制,大多数LHCII异构体被保留下来。因此,nyc1突变体比对照叶片叶绿素降解缓慢,呈现持久的绿色,在叶片衰老后期也能观察到大而密的叶绿体基粒[20]。拟南芥中过表达NOL基因导致叶绿素a/b的比率下降,其光系统II(photosystemII,PSII)的捕光天线系统变小[21]。
HCAR基因编码7-羟基叶绿素a还原酶,是叶绿素降解途径中的另一类重要酶。康乃馨花瓣和叶子发育过程中,HCAR的转录水平与叶绿素含量呈正相关[22]。虽拟南芥绿苗时期HCAR基因表达水平较高,但过表达HCAR基因植株叶片黄化速率却加速,相应的hcar突变体在黑暗诱导处理中依然为滞绿表型[23]。植物衰老过程中一个主要的脱脂醇酶类PPH,广泛存在于藻类和陆地植物。PPH主要作用于脱镁叶绿素,对叶绿素没有影响,PPH的缺失导致植物非功能型滞绿。PAO定位在叶绿体中,在叶片衰老和成熟过程中含量增加[24]。脱镁叶绿酸a转变为荧光代谢产物pFCCs主要依赖PAO和RCCR。水稻的基因组中存在2个RCCR同源基因RCCR1和RCCR在衰老水稻叶片中变化显著。RCCR1和PAO缺失的水稻突变体会出现致死的表型[25]。
2.2 叶绿素降解的调控植物叶绿素降解机制研究中,SGR1(STAY-GREEN1)/NYE1(NON-YELLOWING1)的发现是里程碑进展。进一步研究发现,拟南芥中含3个SGR的同源蛋白,分别为SGR1、SGR2和SGR-LIKE(SGRL),在叶绿素降解中各司其职[26]。其中,对SGR1的研究最为广泛。在自然衰老和黑暗诱导情况下,sgr1-1突变体表现出滞绿现象,且SGR1过表达植株叶片提前黄化[16, 27]。nye1-1/sgr1-1突变体在种子褪绿阶段和营养生长期受生物胁迫和非生物胁迫时,叶片颜色更为深绿[28]。SGRL也促进叶绿素的降解,且仅在正常植物营养阶段发挥功能,但在受胁迫时诱导叶片黄化[29]。大部分陆生植物都存在功能保守的SGR1和SGRL,并在叶绿素降解中发挥重要功能[30]。SGR2和SGR1在序列上高度相似,但功能却截然相反。SGR2过表达植株表现为滞绿,而sgr2突变体叶片提前黄化。这些结果表明,SGR2在叶片衰老过程中,对叶绿素的降解起负调控作用[31]。相比于SGR1和SGR2,SGRL在植物衰老前表达较高,随后表达量开始下降。进一步研究发现,SGR同源蛋白功能的差异可能是与CCEs结合能力不同引起。SGR1和SGRL能与6个CCEs和LHCⅡ互作参与叶绿素代谢途径[26]。值得一提的是,SGR1与CCEs的结合仅存在于植物衰老组织中。然而,SGR2却很难结合CCEs,虽然其可以与LHCⅡ发生互作,但对SGR1-CCE和SGRL-CCE与LHCⅡ的结合起到抑制作用(图 1)[27, 29, 31]。
激素信号通路在叶绿素降解调控中发挥重要作用。水杨酸(salicylic,SA)途径中,WRKY53主要在植物衰老的早期阶段表达,而将其过量表达和敲除后植物分别出现早衰和滞绿表型[33, 34],并且在水杨酸的诱导下,WRKY53作为转录因子激活SAG12(Senescence-associated gene 12)、CATLASE1/2/3和ORE9(ORESARA9)[35]促进植物叶片衰老,影响植物叶绿素的降解。2007年研究发现,EPITHIOSPE-CIFYING SENESCENCE REGULATOR(ESR)随叶片年龄逐渐减少,能够和WRKY53互作入核,并抑制WRKY53的DNA结合功能,以调节植物衰老过程的叶绿素降解[36]。脱落酸(abscisic acid,ABA)通路两个bZIP家族的转录因子ABA INSENSITIVE5(ABI5)和ENHANCED EM LEVEL(EEL)在植物衰老阶段直接激活SGR、NYC1和NYE1,并调控其下游基因PHYTOCHROME INTERACTING FACTOR 4(PIF4)和PIF5的表达[37]。NAC016是叶片衰老相关转录因子,NAC016过表达植株加速衰老,而敲除突变体nac016则衰老延后[38]。通过酵母单杂交和染色质免疫共沉淀(chromatin Immunoprecipitation,ChIP)实验发现,NAC016与SGR1启动子直接结合,调控叶绿素降解[39]。此外,NAC016通过间接方式调控ABA信号通路中的ABI5、EEL和AAO3(ABS-CISIC ALDEHYDE OXIDASE3)表达,影响叶绿素降解。而NAP(NAC-LIKE,ACTIVATED BY AP3/PI)蛋白可以直接结合到ABA合成基因AAO3启动子上,增加植物体内ABA含量来促进SGR和CCEs的含量调控植物衰老[40]。
乙烯(ethylene)可以催化叶绿素的降解[41-43]。乙烯信号通路重要转录因子ETHYLENE INSENSITIVE3(EIN3)通过结合NYC1、NYE1和PAO三个叶绿素代谢基因直接促进叶绿素的降解;ORESARA1(ORE1/NAC092)属于NAC家族并为miR164的靶基因,通过调控一系列涉及到植物信号传导,衰老相关基因SAGs(SENSECESENCE-ASSOCIATED GENES)影响叶片衰老[44, 45]。NAC家族基因ANAC046调控NYC1、SGR1、SGR2和PAO的表达[46]。进一步研究发现,EIN3还可以直接结合到miR164a启动子上抑制其表达,从而促进ORE1基因表达。而ORE1又能在衰老阶段调控乙烯合成基因1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE 2(ACS2),调节乙烯的合成[47]。
植物中茉莉酸(jasmonate,JA)也能诱导植物的衰老。Schommer等[48]发现miR319靶基因TCPs(TEOSINTE BRANCHED1、CYCLOIDEA和PCF)通过转录激活茉莉酸合成基因LIPOXYGENASE 2(LOX2)正向调控叶片衰老。MYC2/3/4蛋白作为茉莉酸信号途径的重要元件,可以直接与CCGs(Chl catabolic enzyme genes)(包括PAO、NYC1和NYE1)基因结合,调控叶绿素降解。此外,MYC2蛋白还能与ANAC019直接互作,调控CCGs(NYE1/SGR1、NYE2/SGR2和NYC1)表达来影响叶绿素降解[47]。
油菜素内酯(brassinosteroid,BR)在植物叶绿素降解中发挥了重要作用。WRKY53同时参与调控水杨酸与BR信号通路中,通过调节ORE9表达,促进BRASSINAZOLE-RESISTANT1(BZR1)和BRI1-EMS-SUPPRESSOR1(BES1)表达调控叶绿素降解[49, 50]。BZR1和BES1还可以直接抑制衰老相关的NAC家族基因(NAC002、NAC019、NAC055和NAC072)[51-53]。另外,BZR1激活转录抑制子WRKY57从而抑制SAG4和SAG12基因表达,以阻止叶片提前衰老[54]。
2.3 其他调控叶绿素降解因素1-甲基环丙烯作为一种乙烯抑制剂(1-MCP),处理植物果皮后,叶绿素代谢途径中部分相关基因被抑制表达,而叶绿素酶和红色叶绿素代谢产物还原酶活性不发生改变,但叶绿素降速率减缓[55]。光照也会加速植物叶绿素的降解[56, 57],但植物油脂可以保护植物叶绿素不被光降解[58]。添糖处理后,植物叶绿素降解速率加快,而花青素积累[59]。
盐胁迫时,植物体内5-氨基酮戊酸(ALA)的合成受到抑制,植物体内叶绿素含量降低,而在降解阶段低盐胁迫会提高叶绿素酶的活性,高盐胁迫后会抑制叶绿素酶的活性,加速叶绿素降解[60]。在低温条件下,一氧化碳会降低植物体内叶绿素酶和镁离子螯合酶的活性,防止生物膜的损坏[61]。干旱条件下,植物叶绿素降解速率加快,且适当胁迫处理有利于植物生物量和产量的提高[62]。
3 滞绿滞绿与作物产量正相关,是作物高产的重要指标之一。如滞绿玉米(Zea mays L.)叶面积增大,对光的捕获能力增强,提高了作物的代谢水平,使作物高产[63]。基于滞绿的重要性且和叶绿素代谢密切关联,通过挖掘叶绿素代谢途径突变体,迄今为止,已经鉴定出多个滞绿表型的突变体(表 1)[64]。
基于衰老过程中叶片滞绿的特性,将滞绿分成5种类型: 功能型滞绿A型和B型,非功能型滞绿C型、D型和E型[7]。A型相比于野生型叶绿素降解起始延后,但降解速率不发生改变,如玉米突变体fs854[65],基因型为GGd1d1d2d2的大豆突变体等[66]。B型降解的叶绿素起始时间与野生型相同,而光合速率和叶绿素降解的速率比野生型缓慢,如高粱突变体和烟草突变体g28[67]。C类型的滞绿,在衰老阶段,某段时间叶绿素降解受到抑制,光合速率和叶绿素降解起始与野生型相同,如草地羊毛突变体bf993[68]。D型为植物受到外界胁迫后,植物组织受到破坏,植物体内的叶绿素不发生降解,导致叶绿素永久停留在植物体内。E型为叶绿素降解速率和光合速率与野生型相同,而叶绿素在植物成熟期阶段含量更高,叶绿素降解需要更多时间。因此,功能型滞绿比非功能型滞绿的滞绿表型和光合特性维持更长[7]。
3.2 滞绿突变体滞绿突变体分为两大类,包括能维持光合能力的功能型滞绿突变体和无光合能力的非功能型滞绿突变体。有功能型的滞绿突变体主要表现为衰老起始延迟或衰老过程延缓。在衰老阶段,水稻‘SNU-SG1’剑叶叶绿素含量比对照高,叶片光合速率也更强,植株结实率提高,水稻产量明显提高[69]。拟南芥ore1、ore3和ore9突变体也是功能型滞绿,3种同源基因通过影响植株中的脱落酸、乙烯和甲基茉莉酸的含量影响叶绿素降解。功能型的滞绿能有效提高作物产量。到目前为止,玉米[70]和小麦[71]中鉴定出的功能性滞绿突变体已经应用于生产。
非功能型滞绿突变体的研究也较多。sgr为水稻非功能型滞绿突变体,仅在灌浆期叶片出现滞绿,但叶片光合作用在植物的衰老阶段正常减少。Hiroshi等[72]在水稻中发现NYC4基因,为拟南芥THYLAKOID FORMATION1(THF1)的同源基因,它的突变呈现非功能型滞绿的表型。但拟南芥thf1并无滞绿表型,仅在衰老阶段调控叶绿素蛋白复合体的降解。研究发现,水稻nyc4与sgr突变体都滞绿,但衰老阶段保存叶绿体蛋白的模式却截然不同,特别是D1和D2蛋白[73],表明NYC4在叶绿素蛋白复合体降解功能上区别于SGR[74]。此外,还发现了多个非功能性滞绿突变体,如草田羊毛senescence-induced deficiency(sid)[75]突变体,豌豆cytG和d1d2突变体[76]及拟南芥ore10 ore11[77]等。拟南芥nyc1和nol的植株中叶绿素b大量残留,以维持补光复合物LHCⅡ的稳定,使植物叶绿素不被降解,但是否具有功能尚未确定[18]。在衰老叶片中,非黄化突变体sid脱植基叶绿素a和脱镁叶绿素a显著上升,但没有PAO活性[78]。拟南芥ACCELERATED CELL DEATH1(ACD1)的缺失[79]导致PAO缺陷突变,使成熟叶片脱镁叶绿酸a积累,光依赖和年龄依赖细胞死亡[80]。玉米中PaO缺失突变体letal leaf spot-1(lls1),叶片会持续出现坏死的斑点直到枯萎[81, 82]。而拟南芥acd2突变体成熟叶片也存在斑点并死亡的特点。
4 展望研究证实,叶绿素降解与作物产量密切相关[83, 84]。通过延迟作物叶绿素降解,延长整个生育后期的光合能力,有利于提高干物质合成和输送,有助于作物产量的提高。叶绿素代谢相关研究已持续数十年,从最初的生理化学分析、物理结构研究到近年来的分子水平研究[85],已对高等植物中的叶绿素含量变化,降解的基因调控,及功能应用等有了比较全面的认识。但随着研究进一步的深入,仍有许多问题待阐明,特别是分子机制的研究。如SGRs在叶绿素降解代谢中的确切功能也尚待进一步澄清;小分子物质、激素、生物与非生物胁迫等是通过何种途径直接调控叶绿素降解的;调控叶绿素降解的各种信号途径间是否存在互作;叶绿素的适时降解是如何响应植物发育调控的;叶绿素降解影响农作物产量的具体分子机制是什么等一系列问题有待深入解析。随着滞绿突变体研究的深入,其应用价值也将被不断挖掘,将会在农产品产量和抗性的提高及农产品的储存和运输方面得到应用。而对滞绿突变体的理论研究,将为进一步研究叶绿素的代谢机制、光合作用、生理生化变化、植物衰老机理研究提供新的思路和方向。
| [1] | Von Wettstein D, Gough S, Kannangara CG. Chlorophyll biosynthe-sis. The Plant Cell , 1995, 7 (7) : 1039–1057. DOI:10.1105/tpc.7.7.1039 |
| [2] | Pru?inská A, Tanner G, Anders I, et al. Chlorophyll breakdown:pheophorbide a oxygenase is a rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci USA , 2003, 100 (25) : 15259–15264. DOI:10.1073/pnas.2036571100 |
| [3] | Rüdiger W. Chlorophyll metabolism:from outer space down to the molecular level. Phytochemistry , 1997, 46 (7) : 1151–1167. DOI:10.1016/S0031-9422(97)80003-9 |
| [4] | Benedetti CE, Costa CL, Turcinelli SR, et al. Differential expression of a novel gene in response to coronatine, methyl jasmonate, and wounding in the Coi1 mutant of Arabidopsis. Plant Physiology , 1998, 116 (3) : 1037–1042. DOI:10.1104/pp.116.3.1037 |
| [5] | Gräfe S, Saluz HP, Grimm B, et al. Mg-chelatase of tobacco:the role of the subunit CHL D in the chelation step of protoporphyrin IX. Proc Natl Acad Sci USA , 1999, 96 (5) : 1941–1946. DOI:10.1073/pnas.96.5.1941 |
| [6] | Hörtensteiner S. Chlorophyll degradation during senescence. Annual Review of Plant Biology , 2006, 57 : 55–77. DOI:10.1146/annurev.arplant.57.032905.105212 |
| [7] | Thomas H, Howarth CJ. Five ways to stay green. Journal of Experimental Botany , 2000, 51 (Suppl 1) : 329–337. |
| [8] | Kräutler B, Jaun B, Matile P, et al. On the enigma of chlorophyll degradation:the constitution of a secoporphinoid catabolite. Angew Chem Int Ed Engl , 1991, 30 (10) : 1315–1318. DOI:10.1002/(ISSN)1521-3773 |
| [9] | Hörtensteiner S, Kräutler B. Chlorophyll breakdown in higher plants. Biochim Biophys Acta , 2011, 1807 (8) : 977–988. DOI:10.1016/j.bbabio.2010.12.007 |
| [10] | Hörtensteiner S. Update on the biochemistry of chlorophyll breakdown. Plant Molecular Biology , 2013, 82 (6) : 505–517. DOI:10.1007/s11103-012-9940-z |
| [11] | Sato Y, Morita R, Katsuma S, et al. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. The Plant Journal , 2009, 57 (1) : 120–131. DOI:10.1111/tpj.2008.57.issue-1 |
| [12] | Meguro M, Ito H, Takabayashi A, et al. Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. The Plant Cell , 2011, 23 (9) : 3442–3453. DOI:10.1105/tpc.111.089714 |
| [13] | Schelbert S, Aubry S, Burla B, et al. Pheophytin pheophorbide hydrolase(pheophytinase)is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. The Plant Cell , 2009, 21 (3) : 767–785. DOI:10.1105/tpc.108.064089 |
| [14] | Rodoni S, Schellenberg M, Matile P. Chlorophyll breakdown in se-nescing barley leaves as correlated with phaeophorbidea oxygenase activity. J Plant Physiol , 1998, 152 (2) : 139–144. |
| [15] | Wüthrich KL, Bovet L, Hunziker PE, et al. Molecular cloning, functional expression and characterisation of RCC reductase involved in chlorophyll catabolism. The Plant J , 2000, 21 (2) : 189–198. DOI:10.1046/j.1365-313x.2000.00667.x |
| [16] | Ren G, An K, Liao Y, et al. Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiology , 2007, 144 (3) : 1429–1441. DOI:10.1104/pp.107.100172 |
| [17] | Ren G, Zhou Q, Wu S, et al. Reverse genetic identification of CRN1 and its distinctive role in chlorophyll degradation in Arabidopsis. Journal of Integrative Plant Biology , 2010, 52 (5) : 496–504. |
| [18] | Horie Y, Ito H, Kusaba M, et al. Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. Journal of Biological Chemistry , 2009, 284 (26) : 17449–17456. DOI:10.1074/jbc.M109.008912 |
| [19] | Christ B, Schelbert S, Aubry S, et al. MES16, a member of the methylesterase protein family, specifically demethylates fluorescent chlorophyll catabolites during chlorophyll breakdown in Arabidopsis. Plant Physiology , 2012, 158 (2) : 628–641. DOI:10.1104/pp.111.188870 |
| [20] | Kusaba M, Ito H, Morita R, et al. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. The Plant Cell , 2007, 19 (4) : 1362–1375. DOI:10.1105/tpc.106.042911 |
| [21] | Jia T, Ito H, Tanaka A. The chlorophyll b reductase NOL participa-tes in regulating the antenna size of photosystem II in Arabidopsis thaliana. Procedia Chem , 2015, 14 : 422–427. DOI:10.1016/j.proche.2015.03.057 |
| [22] | Ohmiya A, Hirashima M, Yagi M, et al. Identification of genes associated with chlorophyll accumulation in flower petals. PLoS One , 2014, 9 (12) : e113738. DOI:10.1371/journal.pone.0113738 |
| [23] | Sakuraba Y, Kim YS, Yoo SC, et al. 7-Hydroxymethyl chlorophyll a reductase functions in metabolic channeling of chlorophyll breakdown intermediates during leaf senescence. Biochem Biophys Res Commun , 2013, 430 (1) : 32–37. DOI:10.1016/j.bbrc.2012.11.050 |
| [24] | Yang X, Zhang Z, Joyce D, et al. Characterization of chlorophyll degradation in banana and plantain during ripening at high temperature. Food Chemistry , 2009, 114 (2) : 383–390. DOI:10.1016/j.foodchem.2008.06.006 |
| [25] | Tang Y, Li M, Chen Y, et al. Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. Journal of Plant Physiology , 2011, 168 (16) : 1952–1959. DOI:10.1016/j.jplph.2011.05.026 |
| [26] | Sakuraba Y, Park SY, Paek NC. The divergent roles of STAYGREEN(SGR)homologs in chlorophyll degradation. Molecules and Cells , 2015, 38 (5) : 390–395. DOI:10.14348/molcells.2015.0039 |
| [27] | Sakuraba Y, Schelbert S, Park SY, et al. STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. The Plant Cell , 2012, 24 (2) : 507–518. DOI:10.1105/tpc.111.089474 |
| [28] | Mecey C, Hauck P, Trapp M, et al. A critical role of STAYGREEN/Mendel’s I locus in controlling disease symptom development during Pseudomonas syringae pv tomato infection of Arabidopsis. Plant Physiology , 2011, 157 (4) : 1965–1974. DOI:10.1104/pp.111.181826 |
| [29] | Sakuraba Y, Kim D, Kim YS, et al. Arabidopsis STAYGREEN-LIKE(SGRL)promotes abiotic stress-induced leaf yellowing during vegetative growth. FEBS Letters , 2014, 588 (21) : 3830–3837. DOI:10.1016/j.febslet.2014.09.018 |
| [30] | Barry CS, McQuinn RP, Chung MY, et al. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiology , 2008, 147 (1) : 179–187. DOI:10.1104/pp.108.118430 |
| [31] | Sakuraba Y, Park SY, Kim YS, et al. Arabidopsis STAY-GREEN2 is a negative regulator of chlorophyll degradation during leaf senescence. Molecular Plant , 2014, 7 (8) : 1288–1302. DOI:10.1093/mp/ssu045 |
| [32] | Balazadeh S. Stay-green not always stays green. Molecular Plant , 2014, 7 (8) : 1264–1266. DOI:10.1093/mp/ssu076 |
| [33] | Miao Y, Laun T, Zimmermann P, et al. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Molecular Biology , 2004, 55 (6) : 853–867. DOI:10.1007/s11103-005-2142-1 |
| [34] | Hinderhofer K, Zentgraf U. Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta , 2001, 213 (3) : 469–473. DOI:10.1007/s004250000512 |
| [35] | Xie Y, Huhn K, Brandt R, et al. REVOLUTA and WRKY53 connect early and late leaf development in Arabidopsis. Development , 2014, 141 (24) : 4772–4783. DOI:10.1242/dev.117689 |
| [36] | Miao Y, Zentgraf U. The antagonist function of Arabidopsis WRK-Y53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. The Plant Cell , 2007, 19 (3) : 819–830. DOI:10.1105/tpc.106.042705 |
| [37] | Sakuraba Y, Jeong J, Kang M, et al. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nature Communications , 2014, 5 : 4636. |
| [38] | Kim Y, Sakuraba Y, Han S, et al. Mutation of the Arabidopsis NAC-016 transcription factor delays leaf senescence. Plant and Cell Physiology , 2013, 54 (10) : 1660–1672. DOI:10.1093/pcp/pct113 |
| [39] | Sakuraba Y, Kim YS, Han SH, et al. The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. The Plant Cell , 2015, 27 (6) : 1771–1787. DOI:10.1105/tpc.15.00222 |
| [40] | Liang C, Wang Y, Zhu Y, et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci USA , 2014, 111 (27) : 10013–10018. DOI:10.1073/pnas.1321568111 |
| [41] | Charoenchongsuk N, Ikeda K, Itai A, et al. Comparison of the expression of chlorophyll-degradation-related genes during ripening between stay-green and yellow-pear cultivars. Scientia Horticulturae , 2015, 181 : 89–94. DOI:10.1016/j.scienta.2014.10.005 |
| [42] | Cheng Y, Guan J. Involvement of pheophytinase in ethylene-mediated chlorophyll degradation in the peel of harvested ‘Yali’ pear. J Plant Growth Regul , 2014, 33 (2) : 364–372. DOI:10.1007/s00344-013-9383-z |
| [43] | Yin X, Xie X, Xia X, et al. Involvement of an ethylene response factor in chlorophyll degradation during citrus fruit degreening. The Plant Journal , 2016, 86 (5) : 403–412. DOI:10.1111/tpj.2016.86.issue-5 |
| [44] | Rauf M, Arif M, Dortay H, et al. ORE1 balances leaf senescence against maintenance by antagonizing G2-like-mediated transcription. EMBO Reports , 2013, 14 (4) : 382–388. DOI:10.1038/embor.2013.24 |
| [45] | Balazadeh S, Siddiqui H, Allu AD, et al. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. The Plant Journal , 2010, 62 (2) : 250–264. DOI:10.1111/j.1365-313X.2010.04151.x |
| [46] | Oda YC, Mitsuda N, Sakamoto S, et al. The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Scientific Reports , 2016, 6 : 23609. DOI:10.1038/srep23609 |
| [47] | Qiu K, Li Z, Yang Z, et al. EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genetics , 2015, 11 (7) : e1005399. DOI:10.1371/journal.pgen.1005399 |
| [48] | Schommer C, Palatnik JF, Aggarwal P, et al. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biology , 2008, 6 (9) : e230. DOI:10.1371/journal.pbio.0060230 |
| [49] | Wang Y, Sun S, Zhu W, et al. Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Developmental Cell , 2013, 27 (6) : 681–688. DOI:10.1016/j.devcel.2013.11.010 |
| [50] | Yin Y, Wang Z, Mora-Garcia S, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell , 2002, 109 (2) : 181–191. DOI:10.1016/S0092-8674(02)00721-3 |
| [51] | Bajguz A, Hayat S. Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiology and Biochemistry , 2009, 47 (1) : 1–8. DOI:10.1016/j.plaphy.2008.10.002 |
| [52] | Sun Y, Fan XY, Cao DM, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Developmental Cell , 2010, 19 (5) : 765–777. DOI:10.1016/j.devcel.2010.10.010 |
| [53] | Yu X, Li L, Zola J, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J , 2011, 65 (4) : 634–646. DOI:10.1111/tpj.2011.65.issue-4 |
| [54] | Jiang Y, Liang G, Yang S, et al. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid-and auxin-mediated signaling in jasmonic acid-induced leaf senescence. The Plant Cell , 2014, 26 (1) : 230–245. DOI:10.1105/tpc.113.117838 |
| [55] | Cheng Y, Dong Y, Yan H, et al. Effects of 1-MCP on chlorophyll degradation pathway-associated genes expression and chloroplast ultrastructure during the peel yellowing of Chinese pear fruits in storage. Food Chemistry , 2012, 135 (2) : 415–422. DOI:10.1016/j.foodchem.2012.05.017 |
| [56] | Kudoh H, Sonoike K. Irreversible damage to photosystem I by chilling in the light:cause of the degradation of chlorophyll after returning to normal growth temperature. Planta , 2002, 215 (4) : 541–548. DOI:10.1007/s00425-002-0790-9 |
| [57] | Cheng D, Zhang Z, Sun X, et al. Photoinhibition and photoinhibition-like damage to the photosynthetic apparatus in tobacco leaves induced by Pseudomonas syringae pv. Tabaci under light and dark conditions. BMC Plant Biology , 2016, 16 (1) : 1–11. DOI:10.1186/s12870-015-0700-5 |
| [58] | Lee E, Ahn H, Choe E. Effects of light and lipids on chlorophyll degradation. Food Science and Biotechnology , 2014, 23 (4) : 1061–1065. DOI:10.1007/s10068-014-0145-x |
| [59] | Momose T, Ozeki Y. Regulatory effect of stems on sucrose-induced chlorophyll degradation and anthocyanin synthesis in Egeria densa leaves. Journal of Plant Research , 2013, 126 (6) : 859–867. DOI:10.1007/s10265-013-0581-3 |
| [60] | Santos CV. Regulation of chlorophyll biosynthesis and degradation by salt stress in sunflower leaves. Scientia Horticulturae , 2004, 103 (1) : 93–99. DOI:10.1016/j.scienta.2004.04.009 |
| [61] | Wang Y, Luo Z, Du R. Nitric oxide delays chlorophyll degradation and enhances antioxidant activity in banana fruits after cold storage. Acta Physiologiae Plantarum , 2015, 37 (4) : 1–10. |
| [62] | Rolando JL, Ramírez DA, Yactayo W, et al. Leaf greenness as a dro-ught tolerance related trait in potato(Solanum tuberosum L. ). Environmental and Experimental Botany , 2015, 110 : 27–35. DOI:10.1016/j.envexpbot.2014.09.006 |
| [63] | Tollenaar M, Ahmadzadeh A, Lee E. Physiological basis of heterosis for grain yield in maize. Crop Science , 2004, 44 (6) : 2086–2094. DOI:10.2135/cropsci2004.2086 |
| [64] | Thomas H, Ougham H. The stay-green trait. Journal of Experimental Botany , 2014, 65 (14) : 3889–3900. DOI:10.1093/jxb/eru037 |
| [65] | Crafts-Brandner SJ, Below FE, Wittenbach VA, et al. Differential senescence of maize hybrids following ear removal:II. Selected leaf. Plant Physiology , 1984, 74 (2) : 360–367. DOI:10.1104/pp.74.2.360 |
| [66] | Guiamét JJ, Teeri JA, Noodén LD. Effects of nuclear and cytoplasmic genes altering chlorophyll loss on gas exchange during monocarpic senescence in soybean. Plant and Cell Physiology , 1990, 31 (8) : 1123–1130. |
| [67] | Crafts Brandner SJ, Leggett JE, Sutton TG, et al. Effect of root system genotype and nitrogen fertility on physiological differences between burley and flue-cured tobacco. I. Single leaf measurements. Crop Science , 1987, 27 (3) : 535–539. DOI:10.2135/cropsci1987.0011183X002700030023x |
| [68] | Davies TG, Thomas H, Thomas BJ, et al. Leaf senescence in a nonyellowing mutant of festuca pratensis:Metabolism of cytochrome f. Plant Physiology , 1990, 93 (2) : 588–595. DOI:10.1104/pp.93.2.588 |
| [69] | Yoo S, Cho S, Zhang H, et al. Quantitative trait loci associated with functional stay-green SNU-SG1 in rice. Molecules and Cells , 2007, 24 (1) : 83–94. |
| [70] | Kim J, Chung K, Woo H. Three positive regulators of leaf senescence in Arabidopsis, ORE1, ORE3 and ORE9, play roles in crosstalk among multiple hormone-mediated senescence pathways. Genes and Genomics , 2011, 33 (4) : 373–381. DOI:10.1007/s13258-011-0044-y |
| [71] | Gong Y, Ji X, Gao J. Grain sink strength related to carbon staying in the leaves of hybrid wheat XN901. Agricultural Sciences in China , 2009, 8 (5) : 546–555. DOI:10.1016/S1671-2927(08)60245-X |
| [72] | Wang Q, Sullivan RW, Kight A, et al. Deletion of the chloroplast-localized Thylakoid Formation1 Gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiology , 2004, 136 (3) : 3594–3604. DOI:10.1104/pp.104.049841 |
| [73] | Fang C, Li C, Li W, et al. Concerted evolution of D1 and D2 to regulate chlorophyll degradation in soybean. The Plant Journal , 2014, 77 (5) : 700–712. DOI:10.1111/tpj.12419 |
| [74] | Yamatani H, Sato Y, Masuda Y, et al. NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll-protein complexes during leaf senescence. The Plant Journal , 2013, 74 (4) : 652–662. DOI:10.1111/tpj.2013.74.issue-4 |
| [75] | Hilditch PI, Thomas H, Thomas BJ, et al. Leaf senescence in a non-yellowing mutant of Festuca pratensis:proteins of photosystem II. Planta , 1989, 177 (2) : 265–272. DOI:10.1007/BF00392815 |
| [76] | Guiamét JJ, Pichersky E, Nooden LD. Mass exodus from senescing soybean chioroplasts. Plant and Cell Physiology , 1999, 40 (9) : 986–992. DOI:10.1093/oxfordjournals.pcp.a029632 |
| [77] | Oh M, Safarova RB, Zulfugarov IS, et al. ORE10, a protein that regulates stay-greenness in Arabidopsis. Korean Journal o Crop Science , 2006, 51 (2) : 143–143. |
| [78] | Pru?inská A, Tanner G, Aubry S, et al. Chlorophyll breakdown in senescent Arabidopsis leaves. characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol , 2005, 139 (1) : 52–63. DOI:10.1104/pp.105.065870 |
| [79] | Greenberg JT, Ausubel FM. Arabidopsis mutants compromised for the control of cellular damage during pathogenesis and aging. The Plant Journal , 1993, 4 (2) : 327–341. DOI:10.1046/j.1365-313X.1993.04020327.x |
| [80] | Tanaka R, Hirashima M, Satoh S, et al. The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a:Inhibition of the pheophorbide a oxygenase activity does not lead to the “Stay-Green” phenotype in Arabidopsis. Plant and Cell Physiology , 2003, 44 (12) : 1266–1274. DOI:10.1093/pcp/pcg172 |
| [81] | Gray J, Janick-Buckner D, Buckner B, et al. Light-dependent death of maize lls1 cells is mediated by mature chloroplasts. Plant Physiology , 2002, 130 (4) : 1894–1907. DOI:10.1104/pp.008441 |
| [82] | Gray J, Close PS, Briggs SP, et al. A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell , 1997, 89 (1) : 25–31. DOI:10.1016/S0092-8674(00)80179-8 |
| [83] | 张恒善, 程砚喜, 王大秋, 等. 大豆结荚期品种间叶绿素含量差异与产量相关分析. 大豆科学 , 2001, 20 (4) : 275–279. |
| [84] | 朱宝国, 张春峰, 王囡囡, 等. 不同追氮方式对寒地玉米相关性状及产量的影响. 核农学报 , 2015, 29 (9) : 1806–1812. |
| [85] | 邢才, 王贵学, 黄俊丽, 等. 植物叶绿素突变体及其分子机理的研究进展. 生物技术通报 , 2008 (5) : 10–12. |