The Cryptromeria fortunei within the Taxodiaceae family are fast-growing evergreen trees. It is one of the main forest plantation species in the subtropical high-altitude areas in China. Taxodiaceae families are relict plants from Cretaceous period. The family was an important component in forest vegetation of the northern hemisphere from the late Cretaceous to the mid-tertiary eras approximately 115-30 million years ago. In the late tertiary and pleistocene, however, the family underwent a widespread reduction resulting in the present-day relictual genera with restricted distributions (Sehlarbaum and Tsuehiya 1984).
At precent, the Cryptromeria is olictypic genera that consists of only 2 species, including C. fortunei and Cryptromeria japonica. The natural distribution of C. fortunei is extremely narrow, consisting of interrupted or isolated distribution in the Wuyishan and Tianmu Mountains in China (Wang et al. 2007). In recent years, driven by economic interests and influenced by environmental factors such as insect damage, C. fortunei resources have sharply fallen. Plant genetic resources are an important material basis for breeding and species diversity protection. Therefore, it is urgent to collect and protect C. fortunei resources. Genetic diversity generally refers to the genetic diversity within a species. The level of genetic diversity directly affects the sustainable development of a population. Molecular markers are an important tool to study genetic diversity. The polymorphisms revealed by molecular markers have no impact by external environment and internal development stage, and therefore are suitable for genetic diversity evaluation. Expressed sequence tag (ESTs) are generated from single-pass sequencing of randomly picked cDNA clones (Adams et al. 1991). The EST approach and subsequent gene-expression profiling (cDNA microarrays) have proven to efficiently evaluate genetic diversity (Enorki et al. 2002, Riar et al. 2011). ESTs are necessary for developing EST-SSR markers.
With the recent progress made in the large-scale function genome sequencing project, thousands of open data sets have been generated. So far, however, there remains no EST data, nor EST-SSR markers, in C. fortunei. According to Yang et al. (2007), EST-derived markers are likely to be conserved across a broader taxonomy than any other sort of marker. For those species with scarce sequence information, it is may be feasible to develop EST-SSR markers using EST data for closely related taxa (Lu et al. 2013). C. japonica, which has lots of sequencing and biological information in public databases, is closely related to C. fortunei (Futamura et al. 2008; Tani et al. 2003, 2004).
Moriguchi et al. (2009) exploited EST-SSR markers in C. japonica and 27 EST-SSR marker sequences have been published. In theory, those markers might be used in genetic diversity analysis in C. fortunei; however, those markers exploited in C. japonica are not enough to meet the need of C. fortunei studies. To meet this need, we provide a study of the utility of freely available C. japonica EST resources for the development of markers necessary for genetic diversity analyses of C. fortunei.
Materials and methods Collection of germplasm resourcesIn the spring of 2013, 49 samples of C. fortunei germplasm were collected from Zhejiang and Fujian provinces, China, respectively (Table 1). The new tender branches of C. fortunei were cut and brought back to the Laoshan forest farm, located in Zhejiang province. The tender branches were grafted cambium to cambium. After 2 months, the shoots made a strong union with the stock and the new leaves emerged.
|
|
Table 1 Collection sites of Cryptromeria fortunei germplasm resources |
The new leaves from C. fortunei were processed for DNA isolation. Total genomic DNA was isolated from 200 mg of fresh leaf using the CTAB method (Murray and Thompson 1980). DNA quality was detected using a UV spectrophotometer and DNA in accordance with the standard was used in following experiments.
Search of putative SSRIn total, 24, 299 EST sequences of C. japonica, released by the plant GDB (http://www.plantgdb.org) were examined. We used simple sequence repeat identification tool (SSRIT) (http://www.gramene.org/db/searches/ssrtool) to search SSR among these ESTs. Those ESTs including a 2-4 bp repeat motifs were selected as putative SSR. To develop candidate SSR markers from the SSRs identified with SSR Finder, we designed PCR primers based on flanking regions on the EST sequences using ePrimer3 (Rozen and Skaletsky 2000). To increase the quality and usability of the silicon exploited SSR markers, we required exact matches between primers and templates, and the product sizes were between 100 and 300 bp.
Verification of SSR markers in C. fortuneiEighty primers were selected and synthesized by Songgong Engineering & Technology Company in China. Three C. fortunei samples, which represent different ecotypes, were used to verify the candidate SSR markers. PCR was performed in 20 μL reactions containing 50 ng of template DNA, 0.5 μmol/L of each primer, 200 μmol/L of each dNTP, 1.5 mmol/L of MgCl2, 1 unit of Taq polymerase, and 2 μL of 10 × PCR reaction buffer.
The following PCR program was used: 5 min at 95 ℃; 30 cycles of 30 s at 95 ℃; 30 s at 56 ℃; 1 min at 72 ℃; and 7 min at 72 ℃ for a final extension. For those primer pairs that did not generate good amplification results, the initial annealing temperatures were adjusted from 55 to 60 ℃. Each of the primer pairs was tested at least twice to confirm the repeatability of the observed bands in each sample. PCR products were separated on 0.8 % agarose gel. Gels were stained with ethidium bromide for visualizing DNA bands.
Diversity analysis of C. fortuneiPrimer pairs that generate good amplification results were used for analysis of the diversity of 49 C. fortunei. PCR reactions were the same as mentioned above. Only clear and reproducible PCR products were separated on 6 % non-denaturing polyacrylamide gel (80 Volts, 2.5 h). Gels were silver stained following the procedure in Xu et al. (2002). Bands were scored as present (1) or absent (0) directly from the gel. Based on the PCR data, we evaluated the allelic diversity of each SSR marker using the polymorphism information content (PIC) value, defined as PICi = 1 -∑Pi2, where Pi is the frequency of the ith marker. POPGENE 32 was used to calculate the observed number of alleles (Na), effective number of alleles (Ne), and Shannon's information index (Ⅰ). Genetic relationships and their grouping on the basis of the genetic distance yielded SSR markers that were assessed using Nei's coefficients (Nei and Li 1979). A dendrogram was generated from the genetic distance matrix using the unweighted pair-group method with arithmetic averages UPGMA, using the NEIGHBOR model of the NTSYS Version 2.1 software (Ueno et al. 2012).
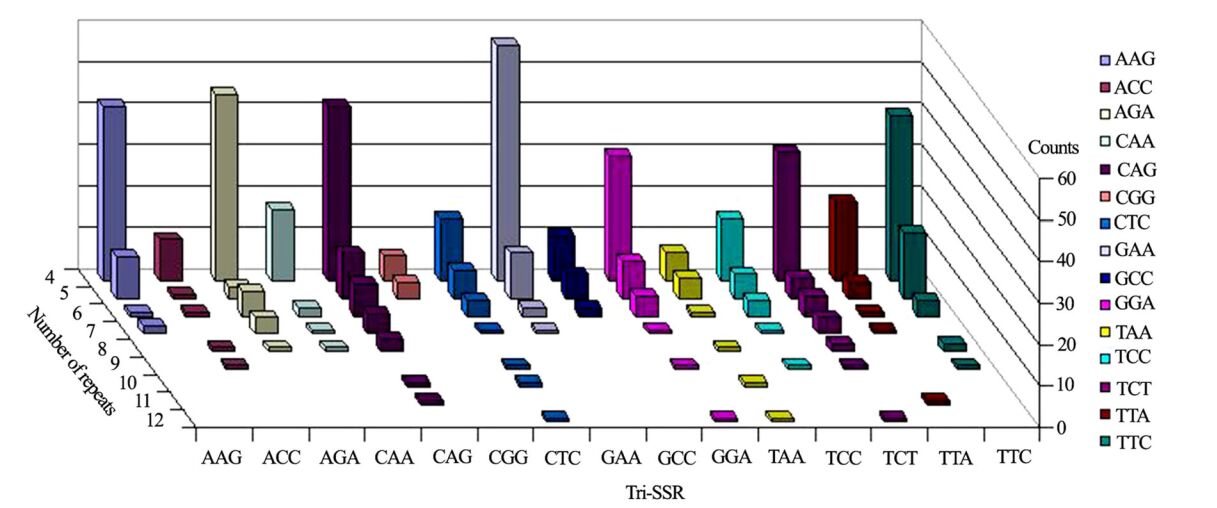
Results and discussion Candidate SSR markersBy screening 24, 299 EST sequences from C. japonica with SSR Finder, we identified 2384 ESTs carrying 2783 SSR motifs. These results showed that 9.8 % ESTs have SSR. Among these ESTs, 2068 contain one SSR motif, 252 contain 2 SSR motifs, 46 contain 3 SSR motifs, 17 contain 4 SSR motifs, and 1 contains 5 SSR motifs. Tri-SSRs were the most common SSRs, counting for 48.47 %, and the most common motif wasGAA(Fig. 1).Thefrequency of the other SSR types and their most common motifs are in Table 2.
|
Fig. 1 Proportions of Tri-SSRs motifs within each type |
|
|
Table 2 SSR number, abundance and distribution frequency of each motif |
We successfully obtained 364 (15 %) primers from 2419 putative SSR loci. Those that did not yield a product were the result of too short sequences flanking the SSRs. The candidate primers were selected and named with the abbreviation CF (for primer C. fortunei) followed by a unique number (e.g., Cf 10). We selected 80 candidate SSR markers for further testing.
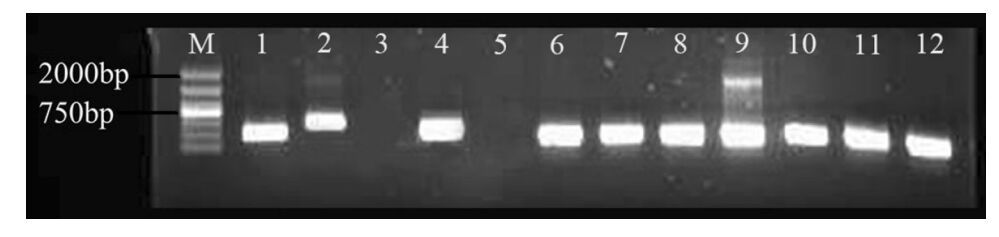
Experimental tests of candidate SSR markersAll primers were tested on three different C. fortunei DNA. Of the 80 candidate SSR markers tested, 70 (87.5 %) yielded stable and clear PCR products (Fig. 2). Because the primers were designed from C. japonica EST data, the result showed the transferability of these two species is relatively high. The primers of these new C. fortunei SSR markers and their name are shown in Table 3.
|
Fig. 2 Candidate primers amplified in C. fortunei |
|
|
Table 3 Candidate SSR primers |
The genetic diversity of 49 C. fortunei was studied. The results showed that 18 primers yielded polymorphism within these accessions. The information of 18 polymorphic primers can be seen in Table 4.
|
|
Table 4 EST-SSR information of 18 polymorphic primers |
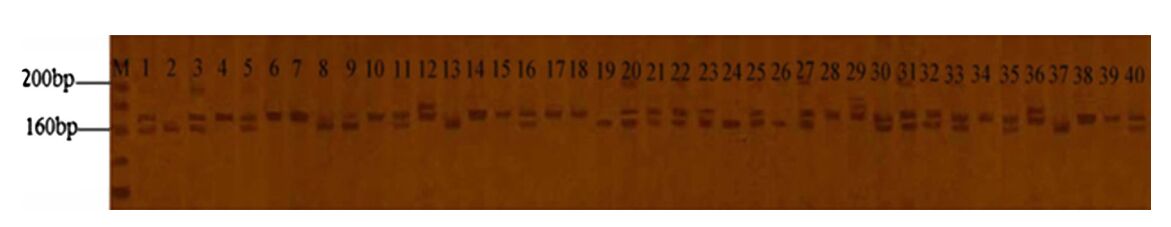
These 18 primers generated 48 scorable SSR loci and the average number of polymorphic SSR loci per primer was 2.7. Patterns of variation observed with the primer CF18 can be seen in Fig. 3. Clearly, SSR variation existed among the 49 C. fortunei. The PIC value varied from 0.375 to 0.8101, with an average of 0.4780. The Shannon index is 0.5718, and the value of Na and Ne are 1.9167 and 1.7289, respectively.
|
Fig. 3 Polymorphisms of C. fortunei amplified by CF18. Note the number correspondence to the resources number in Table 1 |
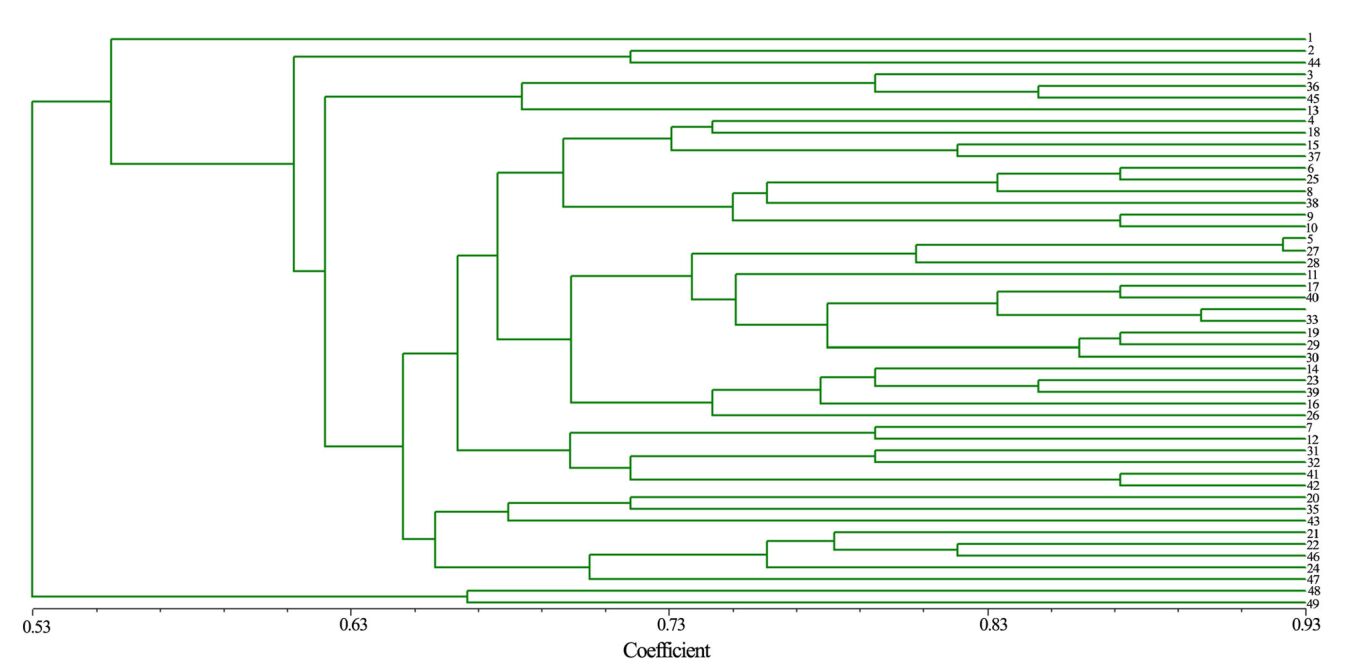
The genetic coefficient of the 49 accessions varied from 0.28 to 0.87. A cluster tree was constructed, according to the genetic distances (Fig. 4). At genetic coefficient of 0.60, the 49 accessions fall into three groups: group 1 contains only FJ-laizhou accession, group Ⅱ contains 2 accessions from FJ-layang, and group 3 contains mixed accessions. It was as expected that 2 accessions from FLlayang formed a group first. Because there is only one accession from FJ-laizhou, determining the genetic distance of this accession is larger than the others. At genetic coefficient of 0.68, the former group Ⅱ was constructed into 7 subgroups, with the first 3 containing 16 accessions in which 11 (69 %) are from FJ province, and the later 4 subgroups containing 31 accessions in which 20 (65 %) were from ZJ province.
|
Fig. 4 Dendrogram of 49 accessions of C. fortunei. Note the number correspondence to the resources number in Table 1 |
EST-SSR genetic markers reside in gene sequences that can directly reflect aspects of variation within those genes. Therefore, the genetic relationship tree constructed with EST-SSR markers could be especially valuable for genetic studies. However, levels of polymorphism could be low because these EST-SSRs are in expressed regions that have more evolutionary conservation compared with primers derived from genome SSR (gSSR) (Xu et al. 2014). However, results from poplar (Zhang et al. 2011) showed that, compared with gSSR, EST-SSR markers were superior in versatility for different species of poplar, and EST-SSR loci revealed higher overall levels of genetic diversity than gSSR loci. At present, however, no EST data remains in C. fortunei. According to Yang et al. (2007), ESTderived markers are likely to be conserved across a broader taxonomic range than any other type of marker. For those species with a scarcity of sequence information, EST data from closely related species could be used. Therefore, we used C. japonica EST data to exploit SSR markers for C. fortunei. On the other hand, our results showed that those sequence data were transferable among these 2 species. Ueno et al. (2012) developed an open framework for the analysis of microsatellites in expressed sequence tags. They used this pipeline to develop EST-SSR markers for C. japonica. However, the SSR frequency detected by their framework was lower (4.54 %) than our method (9.8 %). Consequently, our study developed an economic effectiveness analyzing tool for development of SSR markers in C. fortunei.
C. japonica and C. fortuneiThere has long been argument about classification of the Cryptomeria genus. Most Chinese taxonomists consider Cryptomeria genus to include two species: C. japonica and C. fortunei (namely Chinese Cryptomeria). Some others consider that Chinese C. fortunei is a variety of C. japonica. According to our studies, most primers (70 primers out of 80) yielded from C. japonica EST can be used in C. fortunei, which indicated that the nucleic acid sequence of the two species has higher similarity. But on the other hand, 10 primers out of 80 EST-SSR from C. japonica yielded no amplification products in C. fortunei, suggesting that the two species in the DNA sequence also developed a certain degree of differentiation. The long-term geographical separation may lead to population genetic drift, resulting in the formation of two different populations. Although the difference is relatively small, the DNA sequence differences can be inherited and generate new genotype.
Genetic diversity of C. fortuneiC. japonica is one of the oldest species in China. Using 18 pairs of EST-SSR primers, we detected 49 accessions of C. fortunei. The Shannon index of C. fortunei population that we used in this study is 0.5718, which is relatively high, however large errors may exist. In this study, C. fortunei resources were collected from Zhejiang and Fujian. Some provenances, such as Fujian-Laizhou and Fujian-Layang, have few live C. fortunei trees, so we only able to obtain one or two samples. This may be the main reason for the high Shannon index. We should give more protection to the C. fortunei resources.
In genetic diversity analysis of the 49 resources, a small amount of different geographic origins cluster together, which implies that the genetic diversity of C. fortunei in China is fairly low, probably because the species is windpollinated and long-lived. Takahashi et al. (2005) investigated the allelic variation and genetic structure of C. japonica, and the results showed that no clear relationship between Nei's genetic distances and geographical locations of the populations were found using the principal coordinate and unweighted pair-group methods with arithmetic averaging analyses. In this study, no clearly relationship was found between genetic distances and geographical locations in the C. fortunei group as well. Therefore, during the process of resources conservation and breeding for C. fortunei, the geographical distance, as well as genetic distance should be considered.
Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, Kerlavage AR, McCombie WR, Venter JC (1991) Complementary DNA sequencing: expressed sequence tag and human genome project. Science 252: 1651-1656. DOI:10.1126/science.2047873 |
Enorki H, Sato H, Koinuma K (2002) SSR analysis of genetic diversity among maize inbred lines adapted to cold regions of Japan. Theor Appl Genet 104: 1270-1277. DOI:10.1007/s00122-001-0857-1 |
Felsenstein J (1989) PHILIP-Phylogeny inference package. Cladistics 5: 164-166. |
Futamura N, Totoki Y, Toyoda A, Igasaki T, Nanjo T, Seki M, Sakaki Y, Mari A, Shinozaki K, Shinohara K (2008) Characterization of expressed sequence tags from a full-length enriched cDNA library of Cryptomeria japonica male strobili. BMC Genomics 9: 383-396. DOI:10.1186/1471-2164-9-383 |
Lu Y, Jia Q, Tong Z (2013) Development of amplified consensus genetic markers in Taxodiaceae based on Cryptomeria japonica ESTs data. J For Res 24: 503-508. DOI:10.1007/s11676-013-0381-6 |
Moriguchi Y, Iwata H, Ujino-Ihara T (2003) Development and characterization of microsatellite markers for Cryptomeria japonica D. Don. Theor Appl Genet 106: 751-758
|
Moriguchi Y, Ueno S, Ujino-Ihara T, Futamura N, Matsumoto A, Shinohara K, Tsumura Y (2009) Characterization of EST-SSRs from Cryptomeria japonica. Conserv Genet Resour 1: 373-376. DOI:10.1007/s12686-009-9086-8 |
Murray MG, Thompson WF (1980) Rapid isolation of high molecular-weight plant DNA. Nucl Acids Res 8: 4321-4325. DOI:10.1093/nar/8.19.4321 |
Nei M, Li W (1979) Mathematical model for study the genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci 74: 5267-5273. |
Riar DS, Rustgi S, Burke IC, Gill KS, Yenish JP (2011) EST-SSR development from 5 Lactuca species and their use in studying genetic diversity among L. serriola biotypes. J Hered 102: 17-28. DOI:10.1093/jhered/esq103 |
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365-386. |
Sehlarbaum SE, Tsuehiya T (1984) Cytotaxonomy and phylogeny in certain species of Taxodiaceae. Syst 147: 29-54. |
Takahashi T, Tani N, Taira H, Tsumura Y (2005) Microsatellite markers reveal high allelic variation in natural populations of Cryptomeria japonica near refugial areas of the last glacial period. J Plant Res 118: 83-90. DOI:10.1007/s10265-005-0198-2 |
Tani N, Takahashi T, Iwata H, Mukai Y, Ujino-Ihara T, Matsumoto A, Yoshimura K, Yoshimaru H, Tsumura Y (2003) A consensus linkage map for Sugi (Cryptomeria japonica) from two pedigrees, based on microsatellites and expressed sequence tags. Genetics 165: 1551-1568. |
Tani N, Takahashi T, Ujino-Ihara Y, Iwata H, Yoshimura K, Tsumura Y (2004) Development and characteristics of microsatellite markers for sugi (Cryptomeria japonica D. Don) derived from microsatellite-enriched libraries. Ann For Sci 61: 569-575
|
Ueno S, Moriguchi Y, Uchiyama K, Tsumura Y (2012) A second generation framework for the analysis of microsatellites in expressed sequence tags and the development of EST-SSR markers for a conifer, Cryptomeria japonica. BMC Genomics 13: 136-151. DOI:10.1186/1471-2164-13-136 |
Wang J, Liu J, Huang Y, Yang H (2007) The origin and natural distribution of Cryptomeria. J Sichuan For Sci Technol 28: 92-94. |
Xu S, Tao Y, Yang Z, Chu J (2002) A simple and rapid method used for silver staining and gel preservation. Hereditas 24: 335-336. |
Xu Y, Chen J, Li Y, Hong Z, Wang Y, Zhao Y, Wang X, Shi J (2014) Development of EST-SSR and genomic-SSR in Chinese fir. J Nanjing For Univ 38(1): 9-14. |
Yang L, Jin G, Zhao X, Zheng Y, Xu Z, Wu W (2007) PIP: a database of potential intron polymorphism markers. Bioinformatics 23: 2174-2177. DOI:10.1093/bioinformatics/btm296 |
Zhang Y, Peng C, Li Z, Yang Y, Hu X (2011) Genetic diversity of genomic-SSR and EST-SSR markers in interspecies of Poplar. J Northeast For Univ 39(12): 8-11. |
 2015, Vol. 26
2015, Vol. 26
