文章信息
- 邵丹彤, 魏文强.
- Shao Dantong, Wei Wenqiang.
- 上消化道癌及其癌前病变微生物菌群研究进展
- Progress in research of human microbiota for upper gastrointestinal tumors and precancerous lesions
- 中华流行病学杂志, 2018, 39(3): 382-386
- Chinese Journal of Epidemiology, 2018, 39(3): 382-386
- http://dx.doi.org/10.3760/cma.j.issn.0254-6450.2018.03.025
-
文章历史
收稿日期: 2017-09-11
20世纪90年代,随着人类环境基因组计划(Environmental Genome Project,EGP)的启动与实施,人们对于环境-基因相互作用对健康的影响进行了深入研究,宏基因组学(Metagenomics)技术应运而生。宏基因组学,又称元基因组学、微生物环境基因组学(Microbial environmental genomics),是借助大规模测序,结合生物信息学工具的一项新的分子生物学技术。该技术是以环境样品中全部微生物为研究对象,通过功能基因的筛选和测序分析,对环境中微生物多样性、种群结构、进化关系、功能活性及其与环境之间的关系进行研究的新方法。它避免了对微生物的纯培养,从而有效解决了传统微生物学在检测和鉴定微生物物种方面的局限性[1]。目前,宏基因组学研究已渗透到农业、土壤、海洋等各个领域。第二代高通量测序技术的推广应用,避免了一代测序文库构建过程中利用宿主菌对样品进行克隆而引起的系统偏差,使测序速度、精度、信息丰富程度大幅提升,从而极大地促进了宏基因组学的发展,为医学领域带来巨大变革。
宏基因组测序包括全基因组和16/18S rRNA等扩增测序。16S rRNA基因是细菌染色体上编码rRNA的DNA序列,存在于所有细菌染色体基因组中,其结构、功能在不同细菌中高度保守,具有高信息量及总长度适宜等诸多优势。采用二代测序技术进行16S rRNA可变区测序,可以全面反映微生物群的组成、分布及丰度信息。由于其经济、高效且数据处理比全基因组测序简单,现已被广泛应用于疾病与微生物菌群关系研究中。
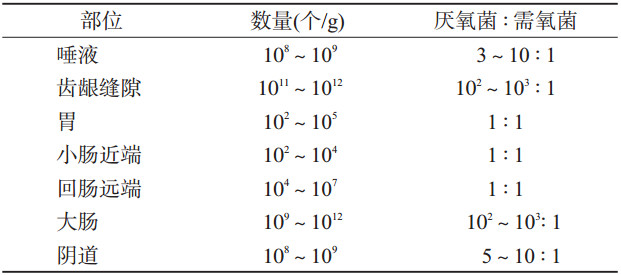
二、微生态环境与机体疾病人体体表、与外界相通腔道及黏膜上定植了大量的微生物群,包括细菌、古生菌、病毒和真菌等,生物分类法以界、门、纲、目、科、属、种对其逐级分类。机体不同部位菌群丰度及构成各异(表 1),其中人体消化道是一个巨大的微生物库,其细胞总量是人体细胞总数的10倍,所含基因数目是人类基因组总和的150倍[2],被称为人体的“第二基因库”。消化道存在大量致病共栖菌谱(Pathobionts),是指导致疾病或健康问题的失衡菌群谱。这些微生物在健康状态下以共生形式存在于肠道,在人体能量代谢[3]、营养吸收[4]、免疫功能[5]和其他生理活动中发挥着重要作用,与宿主共同维持着机体的健康;在非健康状态下其多样性和丰度发生异常变化,导致多种疾病的发生发展,如食管反流、胃炎、伪膜性肠炎[6]等消化道疾病,肥胖[7]、糖尿病等代谢性疾病[8],心血管疾病[9],免疫性疾病[10],帕金森、焦虑、抑郁、自闭症谱系障碍等神经精神性疾病[11-12]及肝癌[13]、结直肠癌[14]、胰腺癌[15]、乳腺癌[16]、黑色素瘤[17]等肿瘤疾病。人群试验表明,粪便移植对于改善90%由于艰难梭菌感染而导致反复腹泻症状具有显著疗效[6];大型出生队列研究显示,为新生儿补充益生菌可降低60%的胰岛自身免疫反应[18]。可见,肠道致病共栖菌与疾病的相关关系已逐步得到证实。关注机体菌群环境,探索疾病与菌群间关联,对于疾病防控、临床诊断和治疗具有重要意义。
2013年,我国新发恶性肿瘤644 487例,死亡399 275例,其中胃癌、肝癌、食管癌和结直肠癌等消化道肿瘤是我国居民发病和死亡的主要负担[19]。目前下消化道肿瘤菌群相关研究较多[20-21],而上消化道肿瘤作为发展中国家主要的肿瘤疾病负担,其与菌群关系的研究尚处于起步阶段。
1.上消化道癌的微生物菌群特征:
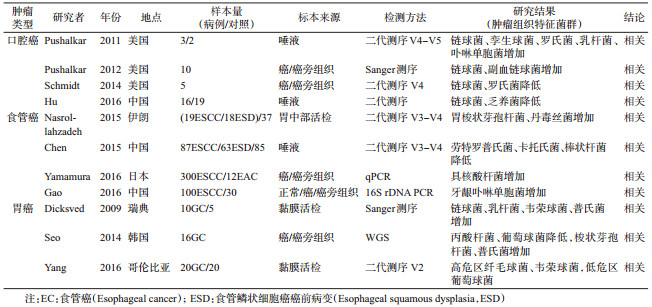
健康口腔黏膜中需氧菌占主导地位,而口腔鳞状细胞癌(Oral squamous cell carcinoma,OSCC)组织病理变化为厌氧菌的定植和生长创造了有利条件,癌组织表面和深部均存在活菌[22]。目前研究表明OSCC患者与健康人口腔菌群存在差异,但菌群变化结论不一[23-26](表 2)。
食管黏膜表面积较大,且位于微生物种类丰富的口咽和胃之间,微生物通过吞咽和反流作用很容易进入食管,进而定植和存活甚至引起疾病。食管癌主要有鳞状细胞癌(Esophageal squamous cell carcinoma,ESCC)和腺状细胞癌(Esophageal adenocarcinoma,EAC)两种病理类型,目前关于食管癌患者菌群的研究甚少且结论不一。Sawada等[27]认为食管癌患者与正常人菌群特征无显著差异,二代测序技术兴起后,某些菌群在食管癌中的特异性表达逐渐被阐明(表 2)。牙龈卟啉单胞菌[28]和具核酸杆菌[29]与ESCC疾病程度和不良预后相关,可将其作为食管癌预后的生物标志物,这两种菌群丰度的升高预示疾病的不良结局。ESCC患者胃部[30]、口腔[31]菌群特征均呈现特异性的改变。EAC患者菌群特征尚不明确,但有多项研究表明Hp(Helicobacter pylori,Hp)对EAC具有保护作用[32-34](表 2),这与Hp抑制胃酸分泌及菌群结构改变有关。
Hp慢性感染是胃癌(Gastric cancer,GC)已经明确的危险因素,Hp阴性与阳性者胃部菌群环境截然不同:Hp阴性者胃部菌群多样性较高,而阳性者菌群主要是Hp,胃癌患者的Hp丰度显著低于正常人[35-36]。但在贲门癌中,Hp相对丰度随着疾病恶性程度的增加而逐步升高,菌群多样性随之下降[37]。除了Hp,胃部还含有庞大而多样的共生菌群系统,胃癌的发生可能有更复杂的菌群参与。动物试验表明,复杂菌群感染较单一Hp感染呈现出更为严重的病理特征[38];一项长达15年的临床试验发现[39],抗Hp治疗者胃癌发病率虽降低,但超过50%的人仍处于Hp感染状态,抗生素治疗可能通过诱导非Hp菌群变化而抑制胃癌的发生。宏基因组学研究深入后,多项研究发现胃癌患者特异性的菌群变化,如乳杆菌、韦荣球菌和普氏菌增多,提示胃癌的发生发展可能与菌群变化存在相关关系。值得注意的是,由于胃部不同解剖结构所处的菌群环境不同,贲门菌群与食管相似,而幽门、胃底和胃体菌群环境更倾向于肠道,后续研究应分别从两个方面深入探讨。
2.上消化道癌前病变的微生物菌群特征:
在亚洲和太平洋地区,咀嚼槟榔是诱发口腔癌前病变及OSCC的首要危险因素,最近Hernandez等[40]发现咀嚼槟榔可通过改变口腔菌群而导致口腔癌前病变。反流性食管炎(Gastroesophageal reflux disease,GERD)和Barrett食管(Barrett’s esophagus,BE)是EAC的癌前病变,不同于正常食管菌群特征,GERD和BE以韦荣球菌、普氏菌、奈瑟菌和弯曲杆菌等革兰阴性菌为主[41-42](表 3),且并非一过性的,这源于革兰阴性菌对于胃酸和胆汁环境的高耐受性。以上菌群中,弯曲杆菌尤其值得引起重视。Blackett等[43]和Macfarlane等[44]发现弯曲杆菌丰度增加的同时其他菌群丰度减少,且IL-8随之上调,提示其可能诱发宿主免疫与炎症反应,这与Hp在胃癌中的变化趋势相似。另外,在正常人群-GERD/BE-EAC进展过程中,菌群丰度显著下降[43],表明菌群数目是随着疾病进展而变化的,但究竟哪些菌的丰度发生改变尚有待于深入研究。
ESCC是包括中国在内的发展中国家食管癌的主要病理类型,关注ESD的菌群变化尤为重要。但目前ESD的菌群研究尤为缺乏,仅有证据表明ESD患者食管菌群丰度降低[45],与GERD/BE菌群特征一致,且口腔菌群由正常-ESD-ESCC逐步降低[31]。目前,中国[46]、印度[47]、伊朗[48]、美国[49]、日本[50]等国家多项研究证实,牙周疾病和牙齿脱落可增加ESD/ESCC的发病风险。考虑到目前食管癌内镜筛查成本高、依从性差等问题,若能识别ESD患者口腔特征菌群,使其作为初筛标志物实现高危人群分流,将大大降低筛查成本并有利于在人群中大规模推广。
与EAC相似,胃癌的菌群结构是随着胃炎-肠上皮化增生-不典型增生-胃癌过程逐步变化的,多项研究均一致表明,在疾病发生发展的过程中乳杆菌[35, 51-53]、毛螺旋菌[51-52]显著升高,关于胃部菌群的多样性结论不一。但现阶段乳杆菌、毛螺旋菌的致病生物学机制尚不明确,需深入探讨其在胃癌的发展中的作用,以期通过对微生物菌群的识别和干预来实现高危人群的分流和胃癌的早期诊断及治疗。
综上所述,基于二代测序技术的宏基因组学可以最大限度地挖掘微生物资源,已经成为国际生命科学技术研究重要的热点和前沿。目前上消化道肿瘤与微生物菌群研究还处于起步阶段,多项流行病学研究均提示上消化道肿瘤与微生物菌群密切相关,但样本量较少且多为病例对照研究,多种潜在因果关联有待前瞻性研究发掘,研究结果尚需大规模人群试验验证。癌前病变的特征性菌群可为肿瘤的早诊早治提供新的理论依据,具有潜在应用前景。识别上消化道癌的标志性菌群不仅可以通过改变菌群组成来治疗疾病,也可用于肿瘤预后复发评估及预测预警。进一步探讨微生物菌群在肿瘤发生发展中的作用及机制,是上消化道肿瘤预防控制领域的研究方向。
利益冲突: 无
| [1] | Huttenhower C, Gevers D, Knight R, et al. Structure, function and diversity of the healthy human microbiome[J]. Nature, 2012, 486(7402): 207–214. DOI:10.1038/nature11234 |
| [2] | Qin JJ, Li RQ, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing[J]. Nature, 2010, 464(7285): 59–65. DOI:10.1038/nature08821 |
| [3] | Backhed F, Ley RE, Sonnenburg JL, et al. Host-bacterial mutualism in the human intestine[J]. Science, 2005, 307(5717): 1915–1920. DOI:10.1126/science.1104816 |
| [4] | Flint HJ, Scott KP, Louis P, et al. The role of the gut microbiota in nutrition and health[J]. Nat Rev Gastroenterol Hepatol, 2012, 9(10): 577–589. DOI:10.1038/nrgastro.2012.156 |
| [5] | Round JL, Lee SM, Li J, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota[J]. Science, 2011, 332(6032): 974–977. DOI:10.1126/science.1206095 |
| [6] | Kassam Z, Lee CH, Yuan YH, et al. Fecal microbiota transplantation for Clostridium difficile infection:systematic review and Meta-analysis[J]. Am J Gastroenterol, 2013, 108(4): 500–508. DOI:10.1038/ajg.2013.59 |
| [7] | Suárez-Zamorano N, Fabbiano S, Chevalier C, et al. Microbiota depletion promotes browning of white adipose tissue and reduces obesity[J]. Nat Med, 2015, 21(12): 1497–1501. DOI:10.1038/nm.3994 |
| [8] | Qin JJ, Li YR, Cai ZM, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes[J]. Nature, 2012, 490(7418): 55–60. DOI:10.1038/nature11450 |
| [9] | Tang WH, Wang ZN, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk[J]. N Engl J Med, 2013, 368(17): 1575–1584. DOI:10.1056/NEJMoa1109400 |
| [10] | Kamada N, Seo SU, Chen GY, et al. Role of the gut microbiota in immunity and inflammatory disease[J]. Nat Rev Immunol, 2013, 13(5): 321–335. DOI:10.1038/nri3430 |
| [11] | Sharon G, Sampson TR, Geschwind DH, et al. The central nervous system and the gut microbiome[J]. Cell, 2016, 167(4): 915–932. DOI:10.1016/j.cell.2016.10.027 |
| [12] | Hsiao EY, Mcbride SW, Hsien S, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders[J]. Cell, 2013, 155(7): 1451–1463. DOI:10.1016/j.cell.2013.11.024 |
| [13] | Dapito DH, Mencin A, Gwak GY, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4[J]. Cancer Cell, 2012, 21(4): 504–516. DOI:10.1016/j.ccr.2012.02.007 |
| [14] | Man SM, Zhu QF, Zhu LQ, et al. Critical role for the DNA sensor AIM2 in stem cell proliferation and cancer[J]. Cell, 2015, 162(1): 45–58. DOI:10.1016/j.cell.2015.06.001 |
| [15] | Quispe-Tintaya W, Chandra D, Jahangir A, et al. Nontoxic radioactive Listeria(at) is a highly effective therapy against metastatic pancreatic cancer[J]. Proc Natl Acad Sci USA, 2013, 110(21): 8668–8673. DOI:10.1073/pnas.1211287110 |
| [16] | Fuhrman BJ, Feigelson HS, Flores R, et al. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women[J]. J Clin Endocrinol Metab, 2014, 99(12): 4632–4640. DOI:10.1210/jc.2014-2222 |
| [17] | Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy[J]. Science, 2015, 350(6264): 1084–1089. DOI:10.1126/science.aac4255 |
| [18] | Uusitalo U, Liu X, Yang JM, et al. Association of early exposure of probiotics and islet autoimmunity in the TEDDY study[J]. JAMA Pediatr, 2016, 170(1): 20–28. DOI:10.1001/jamapediatrics.2015.2757 |
| [19] | Chen WQ, Zheng RS, Zhang SW, et al. Cancer incidence and mortality in China in 2013:an analysis based on urbanization level[J]. Chin J Cancer Res, 2017, 29(1): 1–10. DOI:10.21147/j.issn.1000-9604.2017.01.01 |
| [20] | Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer[J]. Gut, 2017, 66(1): 70–78. DOI:10.1136/gutjnl-2015-309800 |
| [21] | Nakatsu G, Li XC, Zhou HK, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis[J]. Nat Commun, 2015, 6: 8727. DOI:10.1038/ncomms9727 |
| [22] | Hooper SJ, Crean SJ, Lewis MA, et al. Viable bacteria present within oral squamous cell carcinoma tissue[J]. J Clin Microbiol, 2006, 44(5): 1719–1725. DOI:10.1128/JCM.44.5.1719-1725.2006 |
| [23] | Pushalkar S, Ji XJ, Li YH, et al. Comparison of oral Microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma[J]. BMC Microbiol, 2012, 12: 144. DOI:10.1186/1471-2180-12-144 |
| [24] | Pushalkar S, Mane SP, Ji XJ, et al. Microbial diversity in saliva of oral squamous cell carcinoma[J]. FEMS Immunol Med Microbiol, 2011, 61(3): 269–277. DOI:10.1111/j.1574-695X.2010.00773.x |
| [25] | Schmidt BL, Kuczynski J, Bhattacharya A, et al. Changes in abundance of oral microbiota associated with oral cancer[J]. PLoS One, 2014, 9(6): e98741. DOI:10.1371/journal.pone.0098741 |
| [26] | Hu XS, Zhang Q, Hua H, et al. Changes in the salivary microbiota of oral leukoplakia and oral cancer[J]. Oral Oncol, 2016, 56: e6–e8. DOI:10.1016/j.oraloncology.2016.03.007 |
| [27] | Sawada A, Fujiwara Y, Nagami Y, et al. Alteration of esophageal microbiome by antibiotic treatment does not affect incidence of rat esophageal adenocarcinoma[J]. Dig Dis Sci, 2016, 61(11): 3161–3168. DOI:10.1007/s10620-016-4263-6 |
| [28] | Gao SG, Li SG, Ma ZK, et al. Presence of Porphyromonas gingivalis in esophagus and its association with the clinicopathological characteristics and survival in patients with esophageal cancer[J]. Infect Agents Cancer, 2016, 11: 3. DOI:10.1186/s13027-016-0049-x |
| [29] | Yamamura K, Baba Y, Nakagawa S, et al. Human microbiome Fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis[J]. Clin Cancer Res, 2016, 22(22): 5574–5581. DOI:10.1158/1078-0432.CCR-16-1786 |
| [30] | Nasrollahzadeh D, Malekzadeh R, Ploner A, et al. Variations of gastric corpus microbiota are associated with early esophageal squamous cell carcinoma and squamous dysplasia[J]. Sci Rep, 2015, 5: 8820. DOI:10.1038/srep08820 |
| [31] | Chen XD, Winckler B, Lu M, et al. Oral Microbiota and risk for esophageal squamous cell carcinoma in a High-Risk area of China[J]. PLoS One, 2015, 10(12): e143603. DOI:10.1371/journal.pone.0143603 |
| [32] | Peek RM Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas[J]. Nat Rev Cancer, 2002, 2(1): 28–37. DOI:10.1038/nrc703 |
| [33] | Abreu MT, Peek RM Jr. Gastrointestinal malignancy and the microbiome[J]. Gastroenterology, 2014, 146(6): 1534–1546.e3. DOI:10.1053/j.gastro.2014.01.001 |
| [34] | Whiteman DC, Parmar P, Fahey P, et al. Association of Helicobacter pylori infection with reduced risk for esophageal cancer is independent of environmental and genetic modifiers[J]. Gastroenterology, 2010, 139(1): 73–83. DOI:10.1053/j.gastro.2010.04.009 |
| [35] | Dicksved J, Lindberg M, Rosenquist M, et al. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls[J]. J Med Microbiol, 2009, 58(4): 509–516. DOI:10.1099/jmm.0.007302-0 |
| [36] | Seo I, Jha BK, Suh S, et al. Microbial profile of the stomach:comparison between normal mucosa and cancer tissue in the same patient[J]. J Bacteriol Virol, 2014, 44(2): 162. DOI:10.4167/jbv.2014.44.2.162 |
| [37] | Yu GQ, Hu N, Wang LM, et al. Gastric microbiota features associated with cancer risk factors and clinical outcomes:A pilot study in gastric cardia cancer patients from Shanxi, China[J]. Int J Cancer, 2017, 141(1): 45–51. DOI:10.1002/ijc.30700 |
| [38] | Lofgren JL, Whary MT, Ge ZM, et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia[J]. Gastroenterology, 2011, 140(1): 210–220.e4. DOI:10.1053/j.gastro.2010.09.048 |
| [39] | Ma JL, Zhang L, Brown LM, et al. Fifteen-year effects of Helicobacter pylori, garlic, and vitamin treatments on gastric cancer incidence and mortality[J]. J Natl Cancer Inst, 2012, 104(6): 488–492. DOI:10.1093/jnci/djs003 |
| [40] | Hernandez BY, Zhu X, Goodman MT, et al. Betel nut chewing, oral premalignant lesions, and the oral microbiome[J]. PLoS One, 2017, 12(2): e172196. DOI:10.1371/journal.pone.0172196 |
| [41] | Yang LY, Lu XH, Nossa CW, et al. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome[J]. Gastroenterology, 2009, 137(2): 588–597. DOI:10.1053/j.gastro.2009.04.046 |
| [42] | Liu N, Ando T, Ishiguro K, et al. Characterization of bacterial biota in the distal esophagus of Japanese patients with reflux esophagitis and Barrett's esophagus[J]. BMC Infect Dis, 2013, 13: 130. DOI:10.1186/1471-2334-13-130 |
| [43] | Blackett KL, Siddhi SS, Cleary S, et al. Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett's and oesophageal carcinoma:association or causality?[J]. Alimentary Pharmacology & Therapeutics, 2013, 37(11): 1084–1092. DOI:10.1111/apt.12317 |
| [44] | Macfarlane S, Furrie E, Macfarlane GT, et al. Microbial colonization of the upper gastrointestinal tract in patients with Barrett's esophagus[J]. Clin Infect Dis, 2007, 45(1): 29–38. DOI:10.1086/518578 |
| [45] | Yu GQ, Gail MH, Shi JX, et al. Association between upper digestive tract microbiota and cancer-predisposing states in the esophagus and stomach[J]. Cancer Epidemiol Biomarkers Prev, 2014, 23(5): 735–741. DOI:10.1158/1055-9965.EPI-13-0855 |
| [46] | Wei WQ, Abnet CC, Lu N, et al. Risk factors for oesophageal squamous dysplasia in adult inhabitants of a high risk region of China[J]. Gut, 2005, 54(6): 759–763. DOI:10.1136/gut.2004.062331 |
| [47] | Dar NA, Islami F, Bhat GA, et al. Poor oral hygiene and risk of esophageal squamous cell carcinoma in Kashmir[J]. Br J Cancer, 2013, 109(5): 1367–1372. DOI:10.1038/bjc.2013.437 |
| [48] | Abnet CC, Kamangar F, Islami F, et al. Tooth loss and lack of regular oral hygiene are associated with higher risk of esophageal squamous cell carcinoma[J]. Cancer Epidemiol Biomarkers Prev, 2008, 17(11): 3062–3068. DOI:10.1158/1055-9965.EPI-08-0558 |
| [49] | Nwizu NN, Marshall JR, Moysich K, et al. Periodontal disease and incident cancer risk among postmenopausal women:results from the women's health initiative observational cohort[J]. Cancer Epidemiol Biomarkers Prev, 2017, 26(8): 1255–1265. DOI:10.1158/1055-9965.EPI-17-0212 |
| [50] | Sato F, Oze I, Kawakita D, et al. Inverse association between toothbrushing and upper aerodigestive tract cancer risk in a Japanese population[J]. Head Neck, 2011, 33(11): 1628–1637. DOI:10.1002/hed.21649 |
| [51] | Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, et al. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer[J]. Sci Rep, 2014, 4: 4202. DOI:10.1038/srep04202 |
| [52] | Wang LL, Zhou JH, Xin YN, et al. Bacterial overgrowth and diversification of microbiota in gastric cancer[J]. Eur J Gastroenterol Hepatol, 2016, 28(3): 261–266. DOI:10.1097/MEG.0000000000000542 |
| [53] | Eun CS, Kim BK, Han DS, et al. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods[J]. Helicobacter, 2014, 19(6): 407–416. DOI:10.1111/hel.12145 |
 2018, Vol. 39
2018, Vol. 39