文章信息
- 付利万, 张美仙, 吴丽君, 高利旺, 米杰.
- Fu Liwan, Zhang Meixian, Wu Lijun, Gao Liwang, Mi Jie.
- 基因-基因间交互作用对学龄儿童腹型肥胖的影响
- Gene-gene interaction on central obesity in school-aged children in China
- 中华流行病学杂志, 2017, 38(7): 883-888
- Chinese journal of Epidemiology, 2017, 38(7): 883-888
- http://dx.doi.org/10.3760/cma.j.issn.0254-6450.2017.07.007
-
文章历史
收稿日期: 2016-12-06
儿童肥胖率的快速增长已成为严重的公共卫生问题。既往研究发现,中国人与欧裔白种人相比更倾向于腹型肥胖,且罹患糖尿病、心血管疾病,或因肥胖死亡的风险也相对更高[1]。肥胖是由环境因素与遗传因素共同作用引起,遗传因素是肥胖产生的内在基础,即使是生活行为和环境改变,最终也是通过基因表达的改变发挥作用[2]。全基因组关联研究(GWAS)已发现并验证了许多与BMI/一般性肥胖相关的基因多态性位点(SNPs)[3-5],我们通过Meta分析也进行过验证[6];由于腹型肥胖相较于一般性肥胖对于健康危害更大[7],这些SNPs与腹型肥胖是否存在关联,目前研究较少,且结果不一[8-10],加之儿童肥胖遗传易感性可能与成年人不同,所以有待进一步在儿童中验证。在已发现的SNPs所在基因中,FTO(fat mass and obesity associated)基因能够影响能量摄入和消耗,并与BMI和肥胖密切相关[11];MC4R(melanocortin 4 receptor)基因是瘦素介导的食欲调节途径中最末端的基因,主要在下丘脑神经细胞中表达,能够调节能量平衡;BDNF(brain-derived neurotrophic factor)是瘦素、胆囊收缩素调节能量代谢平衡的下游信号,对摄食行为、能量消耗、糖脂代谢等有着直接或间接的调控作用;PCSK1(proprotein convertase subtilisin/kexin type 1)基因可激活阿黑皮素(proopiome-lanocortin,POMC),在瘦素信号转导过程中起调节作用[12];SH2B1(SH2B adaptor protein 1)能调节瘦素和胰岛素活性、调节来自食物的能量利用,调节自身体重[13];CSK(c-src tyrosine kinase)基因可能与脂肪细胞分化过程有关[14]。上述6个基因在下丘脑的瘦素-黑皮质素能量平衡系统或/和脂肪细胞的分化过程中发挥重要作用[11-14],其合成的蛋白在下丘脑中高度表达,故推测这些基因相关的SNPs可能与腹型肥胖具有关联,并且在中枢神经系统代谢通路中可能存在交互作用。为此,本研究以“北京市儿童青少年代谢综合征(Beijing Child and Adolescent Metabolic Syndrome,BCAMS)研究”[15]队列人群为基础,分析上述6个基因的SNPs(FTO rs9939609、MC4R rs17782313、BDNF rs6265、PCSK1 rs6235、SH2B1 rs4788102、CSK rs1378942)与儿童腹型肥胖的关联,并探讨SNPs间交互作用对中国学龄儿童腹型肥胖的影响。
对象与方法1.研究对象:源自2004年4-10月开展的BCAMS研究队列人群[15]。该研究调查了北京地区2万余名6~18岁学生的肥胖及相关代谢异常情况。以筛查出的腹型肥胖学生并接受静脉采血者为肥胖组(1 196人),同时按照1 : 2招募非腹型肥胖2 306名学生作为对照组,调查对象合计3 502人。本研究项目和方案得到首都儿科研究所伦理委员会批准,调查对象均由本人或家长签署书面知情同意书。
2.问卷调查:包括学生年龄、性别、业余体育活动等情况。业余体育活动定义为从事体育课外中等强度运动项目(跑、跳、快走、球类、踢毽子、游泳、滑冰/雪、放风筝、健身等),以每天至少运动30 min为基本运算单位,询问每周运动的天数(赋值:1=每天运动;2=≥3 d/周;3=≥1 d/周;4=≥1 d/2周;5=很少运动)。采用自我报告方法收集父母身高和体重数据,计算BMI,以BMI≥28 kg/m2作为判断父母肥胖的标准。其中双亲至少1名肥胖视为有肥胖家族史。
3.体格检查:按标准方法测量腰围,读数精确至0.1 cm,测2次,取平均值[16]。计算腰围身高比(腰围/身高,WHtR)。标准测量法测定身高和体重,计算BMI[15]。青春期评价按男生测量睾丸容积,女生测量乳房发育,采用Tanner5分期法评估青春期发育阶段[17]。
4.基因多态性检测:采用盐析法从外周血白细胞中提取DNA。通过文献查询和专家研讨,筛选并确定6个基因的多态性位点:FTO rs9939609、MC4R rs17782313、BDNF rs6265、PCSK1 rs6235、SH2B1 rs4788102、CSK rs1378942,使用ABI PrismsTM-7900实时荧光定量PCR仪进行分型检测。本人群的基因型检测成功率>98%。对100例随机样本基因型的复测表明错误率<1%。
5.腹型肥胖定义和分组:以腰围为评价指标,将研究对象分为腹型肥胖和非腹型肥胖两组。采用BCAMS基线总人群腰围性别年龄别第90百分位值(P90)判定腹型肥胖[18]。
6.统计学分析:建立Access数据库,进行数据检查和逻辑纠错。采用SPSS 22.0软件对数据进行统计分析。正态分布资料用x±s表示。计数资料组间比较采用χ2检验,计量资料组间比较采用独立样本t检验。基因型和等位基因分布的哈迪-温伯格平衡(Hardy-Weinberg equilibrium,HWE)采用χ2检验。以加性遗传模型(additive genetic model)分析每个SNPs与腹型肥胖的关联,并采用错误发现率(false discovery rate,FDR)方法进行多重检验校正。采用广义多因子降维法(GMDR)[19-20]分析多个SNP的交互作用,进行符号检验和置换检验,并计算各个维度不同因子组合的交叉验证一致性、平衡检验准确度,同时采用多因素logistic回归模型验证最优GMDR模型中基因-基因交互作用效应。基因-基因交互作用验证分析选用相乘模型,进行Cochran-Mantel-Haenszel分层分析,设OR(AB)为A、B两因素的比值比,OR(A)为A因素的比值比,OR(B)为B因素的比值比,若OR(AB)≠OR(A)×OR(B),则这两个因素存在交互作用。性别、年龄、Tanner分期、体力活动和肥胖家族史作为协变量纳入调整。P<0.05为差异有统计学意义。
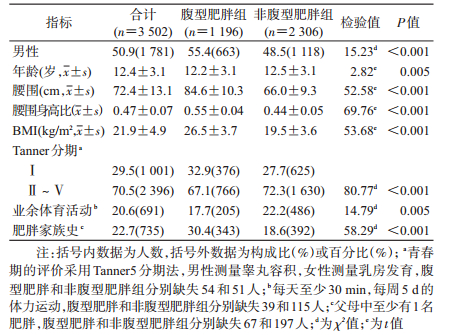
结果1.人群基本特征:3 502名研究对象中,腹型肥胖组男性比例偏高,年龄和业余体育活动偏低,青春发育水平也偏低,肥胖家族史比率高于非腹型肥胖组。腹型肥胖组腰围、WHtR和BMI水平高于非腹型肥胖组(P值均<0.05),见表 1。
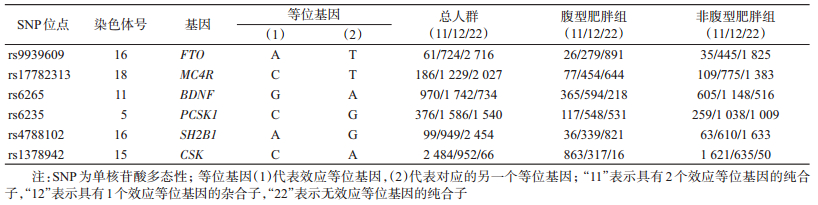
2. HWE检验:6个SNPs(FTO rs9939609、MC4R rs17782313、BDNF rs6265、PCSK1 rs6235、SH2B1 rs4788102和CSK rs1378942)在不同组别的基因型频数分布见表 2。在非腹型肥胖组中各基因型频率的实际值与期望值差异均无统计学意义(P值分别为0.224、0.974、0.521、0.745、0.517、0.160),符合HWE,表明样本来自遗传平衡群体,具有较好的代表性。
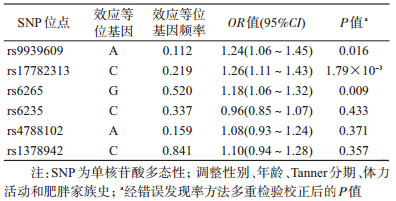
3. SNP位点与腹型肥胖的关联:在调整性别、年龄、Tanner分期、体力活动和肥胖家族史并校正多重检验后,FTO rs9939609-A、MC4R rs17782313-C、BDNF rs6265-G等位基因增加腹型肥胖的罹患风险(P<0.05),而PCSK1 rs6235-C、SH2B1 rs4788102-A、CSK rs1378942-C等位基因与腹型肥胖的关联没有统计学意义(表 3)。
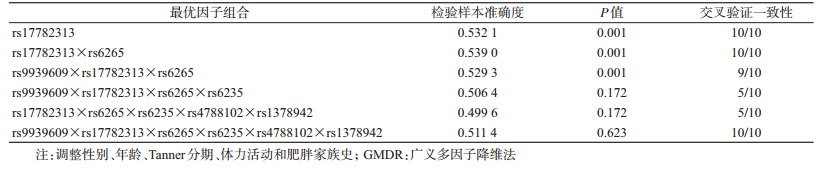
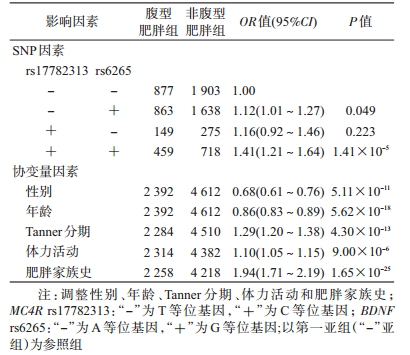
4.基因-基因交互作用与腹型肥胖的关联:以腹型肥胖作为结局变量,将6个SNPs位点纳入GMDR模型作为分析因子,同时将性别、年龄、Tanner分期、体力活动和肥胖家族史作为协变量纳入模型。结果显示,rs17782313和rs6265之间的交互作用有统计学意义(P=0.001),交叉验证一致性为10/10,检验样本准确度为0.539。rs9939609、rs17782313和rs6265之间的交互作用也有统计学意义(P=0.001),交叉验证一致性为10/10,检验样本准确度为0.529 3。其中二阶显著模型的交叉检验一致性和检验样本准确度高于三阶显著模型,据此认为,rs17782313和rs6265两位点的交互作用模型为最佳模型(表 4)。
为进一步验证基因-基因交互作用对腹型肥胖的效应,根据最优GMDR模型组合进行多因素logistic回归分析。以腹型肥胖为因变量,以MC4R rs17782313和BDNF rs6265的等位基因为二分类变量,分别生成哑变量,并且作为分类变量纳入模型方程中,以第一亚组(不含效应等位基因)为参照组,进行MC4R基因和BDNF基因的交互作用logistic回归分析,在调整性别、年龄、Tanner分期、体力活动和肥胖家族史后,结果显示,MC4R rs17782313和BDNF rs6265两位点效应等位基因共同存在时的效应值[OR(AB)=1.41]大于单独存在时效应值的乘积[OR(A)×OR(B)=1.12×1.16=1.30],提示rs17782313与rs6265可能存在正交互作用(表 5)。
本研究结果显示,在调整性别、年龄、Tanner分期、体力活动和肥胖家族史后,FTO rs9939609-A、MC4R rs17782313-C、BDNF rs6265-G等位基因分别独立增加儿童腹型肥胖罹患风险(P<0.05)。既往研究表明,CSK rs1378942位点与中国成年人血压有统计学关联[21]。我们前期研究显示,CSK rs1378942位点与儿童血压无统计学关联[22],而随后经不同体重状态分层后,与肥胖儿童血压存在关联[23]。另外有动物实验发现,CSK基因可能与脂肪细胞的分化有关[14]。因此推测,CSK rs1378942位点也可能与肥胖有关。鉴于CSK rs1378942位点与肥胖的关系目前还未有相关报道,本研究分析了此位点与腹型肥胖的关联,但未发现与儿童腹型肥胖的发生有统计学意义(OR=1.10,95%CI:0.94~1.28,P=0.238)。由于样本来源不同、使用的诊断标准差异、人群的异质性和环境因素的混杂/修饰效应等因素存在,SNP位点与腹型肥胖的关联研究还需要在不同人群中进行不同层次的多方位验证。
肥胖是由多个微效基因和环境因素共同作用的复杂性疾病[24-25],基因的交互作用对肥胖发病的影响不可忽视。2007年Lou等[19]提出的GMDR在2型糖尿病[26]、脑卒中[27]、哮喘[28]、糖尿病肾病[29]等研究领域中成功发现基因-基因交互作用。在病例对照研究中,采用GMDR从研究的总因子组中,通过多重交叉验证评估所有可能的因子组合,评估具有最高预测准确度和/或最高交叉验证一致性的因子组合模型预测疾病状态的能力,并进一步通过置换检验得到最佳因子组合预测模型的统计学意义[30]。而传统logistic回归分析基因-基因交互作用时,因为很多复杂疾病的基因型和表型并非线性关系,所以对模型参数的结果很难解释;另一方面在分析位点数量增加时,所需样本量将呈指数倍增加,由于维度困扰导致logistic回归模型参数估计错误,使效能降低。因此与logistic回归模型等传统参数分析方法相比,GMDR分析集属性选择、构建和分类为一体,不仅对于复杂疾病的高阶交互作用分析和识别有更高的把握度,大大降低建模所需的自由度,同时它的应用不受遗传模式(显性、隐性或加性遗传)和交互作用模型(加法或乘法模型,线性和非线性模型)的限制。不仅如此,GMDR也可用于连续型结局变量分析,能够纳入协变量以提高预测准确率,适用的数据结构也更加宽泛。但是GMDR也存在不足之处,其在分析各因素、各水平交互作用时并不考虑主效应,而logistic回归更能发现主效应,且效应值更加准确[31]。鉴于此,本研究先用GMDR建立模型,最后根据最优GMDR模型结合多因素logistic回归估计危险度,有助于进一步验证基因交互作用对腹型肥胖的效应。GMDR分析结果显示,在调整性别、年龄、Tanner分期、体力活动和肥胖家族史后,1个二阶模型(包括rs17782313和rs6265)和1个三阶模型的交互作用(包括rs9939609、rs17782313和rs6265)有统计学意义,其中二阶模型为最优,应用多因素logistic回归模型验证交互作用效应,调整性别、年龄、Tanner分期、体力活动和肥胖家族史后,研究显示两位点(MC4R rs17782313-C和BDNF rs6265-G)的交互作用可能增加腹型肥胖罹患风险。
本研究存在不足。采用病例对照研究设计,尚无法确定因素与结局之间的因果联系,还需要前瞻性研究加以验证本研究结果;此外,尽管在研究中调整了协变量,如性别、年龄、Tanner分期、体力活动和肥胖家族史后,发现基因-基因交互作用具有统计学意义,但仍不能确定这种交互作用是否由于受其他未知的环境因素混杂影响。
总之,本研究发现FTO rs9939609-A、MC4R rs17782313-C和BDNF rs6265-G等位基因分别独立增加儿童腹型肥胖罹患风险,MC4R rs17782313和BDNF rs6265可能存在交互作用,提示复杂的基因交互作用也可能影响我国学龄儿童腹型肥胖。
利益冲突: 无
| [1] | He W, Zhang S, Song AH, et al. Greater abdominal fat accumulation is associated with higher metabolic risk in Chinese than in white people:an ethnicity study[J]. PLoS One, 2013, 8(3): e58688. DOI:10.1371/journal.pone.0058688 |
| [2] |
赵小元, 张美仙, 程红, 等.
肥胖相关基因多态性对儿童期肥胖发生风险及持续状态的影响[J]. 中华流行病学杂志, 2013, 34(6): 560–565.
Zhao XY, Zhang MX, Cheng H, et al. Risk of obesity-related gene polymorphism on the incidence and durative of childhood obesity[J]. Chin J Epidemiol, 2013, 34(6): 560–565. DOI:10.3760/cma.j.issn.0254-6450.2013.06.005 |
| [3] | Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity[J]. Science, 2007, 316(5826): 889–894. DOI:10.1126/science.1141634 |
| [4] | Loos RJF, Lindgren CM, Li SX, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity[J]. Nat Genet, 2008, 40(6): 768–775. DOI:10.1038/ng.140 |
| [5] | Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity[J]. Nat Genet, 2008, 41(1): 18–24. DOI:10.1038/ng.274 |
| [6] |
付利万, 张美仙, 高利旺, 等.
SEC16B基因多态性与体质指数和肥胖关联的Meta分析[J]. 中华流行病学杂志, 2016, 37(9): 1288–1295.
Fu LW, Zhang MX, Gao LW, et al. Association between SEC16B polymorphisms and body mass index variation or risk of obesity:a Meta-analysis[J]. Chin J Epidemiol, 2016, 37(9): 1288–1295. DOI:10.3760/cma.j.issn.0254-6450.2016.09.021 |
| [7] | Janssen I, Katzmarzyk PT, Ross R. Waist circumference and not body mass index explains obesity-related health risk[J]. Am J Clin Nutr, 2004, 79(3): 379–384. |
| [8] | Chen FF, Wang YF, Shan XY, et al. Association between childhood obesity and metabolic syndrome:evidence from a large sample of Chinese children and adolescents[J]. PLoS One, 2012, 7(10): e47380. DOI:10.1371/journal.pone.0047380 |
| [9] | Tanaka M, Yoshida T, Bin W, et al. FTO, abdominal adiposity, fasting hyperglycemia associated with elevated HbA1c in Japanese middle-aged women[J]. J Atheroscler Thromb, 2012, 19(7): 633–642. DOI:10.5551/jat.11940 |
| [10] | Fawwad A, Siddiqui IA, Basit A, et al. Common variant within the FTO gene, rs9939609, obesity and type 2 diabetes in population of Karachi, Pakistan[J]. Diabetes Metab Syndr, 2016, 10(1): 43–47. DOI:10.1016/j.dsx.2015.02.001 |
| [11] | Zhao X, Yang Y, Sun BF, et al. FTO and obesity:mechanisms of association[J]. Curr Diab Rep, 2014, 14(5): 486. DOI:10.1007/s11892-014-0486-0 |
| [12] | Hebebrand J, Hinney A, Knoll N, et al. Molecular genetic aspects of weight regulation[J]. Dtsch Arztebl Int, 2013, 110(19): 338–344. DOI:10.3238/arztebl.2013.0338 |
| [13] | Doche ME, Bochukova EG, Su HW, et al. Human SH2B1 mutations are associated with maladaptive behaviors and obesity[J]. J Clin Invest, 2012, 122(12): 4732–4736. DOI:10.1172/JCI62696 |
| [14] | Usui M, Uno M, Nishida E. Src family kinases suppress differentiation of brown adipocytes and browning of white adipocytes[J]. Genes Cells, 2016, 21(4): 302–310. DOI:10.1111/gtc.12340 |
| [15] |
米杰, 程红, 侯冬青, 等.
北京市2004年2~18岁儿童青少年超重和肥胖流行现状[J]. 中华流行病学杂志, 2006, 27(6): 469–474.
Mi J, Cheng H, Hou DQ, et al. Prevalence of overweight and obesity among children and adolescents in Beijing in 2004[J]. Chin J Epidemiol, 2006, 27(6): 469–474. DOI:10.3760/j.issn.0254-6450.2006.06.003 |
| [16] | Physical status:the use and interpretation of anthropometry. Report of a WHO Expert Committee[J]. World Health Organ Tech Rep Ser, 1995, 854:1-452. |
| [17] | Marshall WA, Tanner JM. Puberty[M]//Falkner F, Tanner JM. Human growth:a comprehensive treatise. 2nd ed. New York:Plenum Press, 1986:171-210. |
| [18] |
孟玲慧, 米杰.
北京市学龄儿童腰围、腰围身高比分类标准对心血管代谢危险因素的筛查效度[J]. 中国循证儿科杂志, 2008, 3(5): 324–332.
Meng LH, Mi J. The validation of the classification criterion of waist and waist-to-height ratio for cardiometabolic risk factors in Chinese school-age children[J]. Chin J Evid Based Pediatr, 2008, 3(5): 324–332. DOI:10.3969/j.issn.1673-5501.2008.05.002 |
| [19] | Lou XY, Chen GB, Yan L, et al. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence[J]. Am J Hum Genet, 2007, 80(6): 1125–1137. DOI:10.1086/518312 |
| [20] |
陈卿, 唐迅, 胡永华.
应用广义多因子降维法分析数量性状的交互作用[J]. 中华流行病学杂志, 2010, 31(8): 938–941.
Chen Q, Tang X, Hu YH. Detecting interaction for quantitative trait by generalized multifactor dimensionality reduction[J]. Chin J Epidemiol, 2010, 31(8): 938–941. DOI:10.3760/cma.j.issn.0254-6450.2010.08.024 |
| [21] | Li J, Shi JX, Huang W, et al. Variant near FGF5 has stronger effects on blood pressure in Chinese with a higher body mass index[J]. Am J Hypertens, 2015, 28(8): 1031–1037. DOI:10.1093/ajh/hpu263 |
| [22] | Xi B, Shen Y, Zhao X, et al. Association of common variants in/near six genes (ATP2B1, CSK, MTHFR, CYP17A1, STK39 and FGF5) with blood pressure/hypertension risk in Chinese children[J]. J Hum Hypertens, 2014, 28(1): 32–36. DOI:10.1038/jhh.2013.50 |
| [23] | Xi B, Zhao XY, Chandak GR, et al. Influence of obesity on association between genetic variants identified by genome-wide association studies and hypertension risk in Chinese children[J]. Am J Hypertens, 2013, 26(8): 990–996. DOI:10.1093/ajh/hpt046 |
| [24] | Wu LJ, Xi B, Zhang MX, et al. Associations of six single nucleotide polymorphisms in obesity-related genes with BMI and risk of obesity in Chinese children[J]. Diabetes, 2010, 59(12): 3085–3089. DOI:10.2337/db10-0273 |
| [25] | Xi B, Wang CY, Wu LJ, et al. Influence of physical inactivity on associations between single nucleotide polymorphisms and genetic predisposition to childhood obesity[J]. Am J Epidemiol, 2011, 173(11): 1256–1262. DOI:10.1093/aje/kwr008 |
| [26] | Lin E, Pei D, Huang YJ, et al. Gene-gene interactions among genetic variants from obesity candidate genes for nonobese and obese populations in type 2 diabetes[J]. Genet Test Mol Biomarkers, 2009, 13(4): 485–493. DOI:10.1089/gtmb.2008.0145 |
| [27] | Liu JH, Sun K, Bai YY, et al. Association of three-gene interaction among MTHFR, ALOX5AP and NOTCH3 with thrombotic stroke:a multicenter case-control study[J]. Hum Genet, 2009, 125(5/6): 649–656. DOI:10.1007/s00439-009-0659-0 |
| [28] | Chan IHS, Tang NLS, Leung TF, et al. Study of gene-gene interactions for endophenotypic quantitative traits in Chinese asthmatic children[J]. Allergy, 2008, 63(8): 1031–1039. DOI:10.1111/j.1398-9995.2008.01639.x |
| [29] | Wu LSH, Hsieh CH, Pei D, et al. Association and interaction analyses of genetic variants in ADIPOQ, ENPP1, GHSR, PPARγ and TCF7L2 genes for diabetic nephropathy in a Taiwanese population with type 2 diabetes[J]. Nephrol Dial Transplant, 2009, 24(11): 3360–3366. DOI:10.1093/ndt/gfp271 |
| [30] |
李娜, 唐迅, 陈大方, 等.
复杂疾病病因研究中基因间交互作用分析:基于基因型传递不平衡的多因子降维法[J]. 中华流行病学杂志, 2007, 28(10): 1036–1040.
Li N, Tang X, Chen DF, et al. Identification of gene-gene interactions related to the etiology of complex disease:a multifactor dimensionality reduction-genotype pedigree disequilibrium test[J]. Chin J Epidemiol, 2007, 28(10): 1036–1040. DOI:10.3760/j.issn.0254-6450.2007.10.024 |
| [31] |
骆常好, 刘桂芬, 张爱莲.
多因子降维法和logistic回归交互效应分析对比研究[J]. 中国药物与临床, 2008, 8(10): 777–779.
Luo CH, Liu GF, Zhang AL. Analysis of interactions using mulfifactor dimensionality reduction versus logistic regression[J]. Chin Remed Clin, 2008, 8(10): 777–779. DOI:10.3969/j.issn.1671-2560.2008.10.007 |
 2017, Vol. 38
2017, Vol. 38