b School of Public Health, Wuhan University, Wuhan 430071, China;
c Research Center of Public Health, Renmin Hospital of Wuhan University, Wuhan 430060, China;
d Cancer Precision Diagnosis and Treatment and Translational Medicine Hubei Engineering Research Center, Zhongnan Hospital of Wuhan University, Wuhan Research Center for Infectious Diseases and Cancer, Chinese Academy of Medical Sciences, Wuhan 430071, China
Nucleic acids contain a variety of chemical modifications that play critical roles in many biological processes [1-5]. Over 150 different types of modifications have been identified in RNA molecules [6-9]. Adenosine-to-inosine (A-to-I) RNA editing is one of the most important modifications in RNA [10,11]. Inosine (I) is essentially a purine nucleoside resulting from the enzymatic deamination of adenosine (A) at C6 position [12,13]. Inosine was initially identified in the tRNA in yeast [14]. In mRNAs of eukaryotes, inosines are generated by adenosine deaminases acting on RNAs (ADARs) [15].
Inosine preferentially pairs with cytosine, resulting in being recognized as guanine [16]. Meanwhile, inosine can also pair with adenine in a less stable way, thus affecting transcript localization and translation accuracy (Fig. S1 in Supporting information) [16]. Previous studies revealed that inosine was also involved in the incidence and development of many human diseases, including neurological diseases [17], cancers [18] and autoimmune diseases [19]. Uncovering the functions of inosine requires the sensitive and accurate detection of inosine in RNA. The established methods for analysis of nucleic acid modifications include thin-layer chromatography, capillary electrophoresis, mass spectrometry, and sequencing [20-22]. Liquid chromatography−tandem mass spectrometry (LC-MS/MS) has been frequently employed for the quantification of nucleic acid modifications [21,23-27]. However, due to the low ionization of inosine in LC-MS/MS analysis, detection of inosine is challenging, especially in a single cell.
Chemical derivatization strategy can introduce an easily ionizable group into target compounds, which will improve the detection sensitivities of analytes in LC-MS/MS analysis [28,29]. Along this line, here we developed a method by CMCT (N-cyclohexyl-N'-β-(4-methylmorpholinium)ethylcarbodiimide p-toluenesulfonate) derivatization coupled with LC-MS/MS analysis for the sensitive and accurate determination of inosine. Due to the high sensitivity, the developed CMCT derivatization in combination with LC-MS/MS analysis enables the distinct detection of inosine in RNA from a single cell.
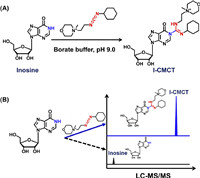
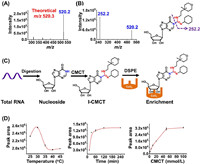
As for the method development, we first examined the CMCT-derivatized inosine (I-CMCT). The carbodiimide group in CMCT can selectively react with the NH group at the N1 position of inosine (Fig. 1A). CMCT contains a positively charged quaternary ammonium group which can be readily ionized and eventually can improve the detection sensitivity of the derivative of I-CMCT in LC-MS/MS analysis (Fig. 1B). As expected, the results showed that the precursor ion (m/z shown in blue) of the detected derivative was identical to its theoretical value (m/z shown in red) (Fig. 2A). Moreover, the product ion (m/z 252.2) of I-CMCT was clearly observed in the product ion MS spectra (Fig. 2B). With the increasing collision energy, we observed the decreased intensity of the precursor ion (m/z 520.2) and the increased intensity of the product ion (m/z 252.2) (Fig. S2 in Supporting information). The precursor ion and the product ion of I-CMCT are different with those of CMCT (Fig. S3 in Supporting information), which excludes the detected ions from CMCT and further confirms the formed derivative of I-CMCT.
|
Download:
|
| Fig. 1. Chemical derivatization reaction between CMCT and inosine. (A) CMCT carries a carbodiimide group that can selectively react with the NH group at the N1 position of inosine. (B) Schematic illustration of the improved detection sensitivity of I-CMCT compared to the native inosine in LC-MS/MS analysis. | |
|
Download:
|
| Fig. 2. Examination and optimization of the CMCT derivatization reaction. (A) Full scan MS spectrum of I-CMCT. (B) Product ion spectrum of I-CMCT. Shown in red and blue are the theoretical m/z and measured m/z, respectively. (C) Schematic illustration of the CMCT derivatization and purification by DSPE with CeO2 to capture I-CMCT and remove excess CMCT. (D) Optimization of the CMCT derivatization reaction conditions. | |
It should be noted that excess CMCT may affect the sensitive detection of I-CMCT in LC-MS/MS analysis. We previously demonstrated that cerium oxide (CeO2) could selectively capture cis-diol compounds under basic condition, and the captured compounds could be released from CeO2 in acidic condition [30]. Since the derivative of I-CMCT carries cis-diol group in the ribose of inosine, we therefore developed dispersive solid phase extraction (DSPE) with using CeO2 to selectively extract I-CMCT, and in the meantime, to remove excess CMCT that contains no cis-diol group (Fig. 2C). The results showed that over 99.7% excess CMCT was removed.
To obtain the best derivatization efficiency, we further optimized the CMCT derivatization conditions, including the reaction temperature, the reaction time and the concentration of CMCT. As for the reaction temperature, it can be seen that reaction at 30 ℃ offered the best reaction efficiency (Fig. 2D). In addition, we observed that the peak area of I-CMCT reached to a plateau at 2 h (Fig. 2D). Moreover, the derivatization efficiency was relatively good when the concentration of CMCT was 50 mmol/L (Fig. 2D). Taken together, the optimized derivatization reaction was carried out at 30 ℃ for 2 h with 50 mmol/L CMCT. By comparing the signal of the residual inosine after CMCT derivatization to the signal of inosine without CMCT derivatization, the derivatization efficiency of the reaction was over 93.4% under the optimized reaction conditions (Fig. S4 in Supporting information). In addition, I-CMCT exhibited good stability (Fig. S5 in Supporting information), indicating the CMCT derivatization is suitable for the analysis of inosine by LC-MS/MS.
We next evaluated the detection sensitivity of inosine after CMCT derivatization under optimized derivatization reactions. In addition, we also optimized the mass spectrometry parameters to achieve the best detection. The detailed information can be found in Tables S1 and S2 (Supporting information). The native inosine has relatively weak retention (1.9 min) on the reversed-phase column (Fig. S4A). However, the retention of I-CMCT was significantly increased (5.1 min) (Fig. 3), which is due to the enhanced hydrophobicity of I-CMCT through the introduction of cyclohexyl group from CMCT. Compared to the native form of inosine, the detection sensitivity of inosine remarkably increased after CMCT derivatization. The limit of detection (LOD), defined as the amounts of the analytes at a signal-to-noise ratio (S/N) of 3, of I-CMCT was 4.5 amol (Table S3 and Fig. S6 in Supporting information). However, the LOD of native inosine was 2.5 fmol (Table S3 in Supporting information). The results demonstrated that the detection sensitivity of inosine improved by 556-fold after CMCT derivatization (Table S3). The significant increase in detection sensitivity of inosine should be attributed to the positive charge of the quaternary ammonium group in the CMCT as well as the increased retention of I-CMCT in chromatographic separation.

|
Download:
|
| Fig. 3. Determination of inosine from RNA. From upper to bottom: the extracted-ion chromatograms of blank control (without addition of RNA), I-CMCT standard, from 300 pg of total RNA, from 300 pg of total RNA with spiked I-CMCT standard, and from 13 pg of total RNA. | |
With the established CMCT derivatization combined with LC-MS/MS analysis method, we then carried out the detection of inosine in RNA from HEK293T cells. The enzymatically digested nucleosides from total RNA of HEK293T cells were reacted with CMCT in borate buffer (pH 9.0), followed by DSPE with CeO2 to capture I-CMCT and remove excess CMCT. After elution with 1% formic acid, the eluent was collected, lyophilized to dryness, and then dissolved in water for LC-MS/MS analysis. The results showed a peak with a retention time of 5.1 min in the extracted-ion chromatogram (m/z 520.2→252.2) from total RNA (300 pg) of HEK293T cells, which was consistent with the retention time of I-CMCT standard (Fig. 3). We also added the I-CMCT standard to the enzymatically digested nucleosides from total RNA of HEK293T cells. The results showed that the spiked I-CMCT standard had the same retention time as that detected in HEK293T cells. Meanwhile, the peak intensity of I-CMCT was increased with the spiked I-CMCT standard (Fig. 3). These results confirmed that the detected compound was inosine. We then further decreased the amount of input RNA, and it can be seen that a clear peak of inosine was observed even from 13 pg of total RNA with the S/N being 26 (Fig. 3). It has been reported that a single mammalian cell contains approximately 10-40 pg of RNA [31,32]. These results indicated that the method should be capable of detection of inosine from a single cell.
We next constructed a calibration curve to quantify inosine level in RNA. In this respect, various amounts of inosine and fixed amount of internal standard (adenosine) were mixed to construct the calibration curve by plotting the mean peak area ratios of I-CMCT/adenosine (I-CMCT/A) versus the mean molar ratios of inosine/adenosine (I/A) based on data obtained from triplicate measurements. The results showed that good linearity was obtained with the coefficient of determination (R2) being greater than 0.99 (Fig. S7 in Supporting information). The relative errors (REs) and intra-day and inter-day relative standard deviations (RSDs) were employed to evaluate the accuracy and precision of the method. As shown in Table S4 (Supporting information), the REs and RSDs were less than 4.3% and 16.7%, respectively. These results showed good precision and accuracy for quantification of inosine, demonstrating that the method was reliable for the accurate quantification of inosine. With the constructed calibration curve, the quantification results showed that the level of inosine from 13 pg of RNA was 0.22% (I/A), which is comparable to the result obtained with using larger amount (300 pg) of RNA (0.28%, I/A).
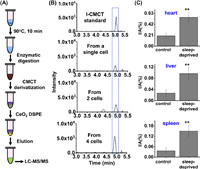
Next, we employed the CMCT derivatization combined with LC-MS/MS analysis method to directly detect inosine in a single cell (Fig. 4A). The samples with a few cells were prepared by successive dilution of large number of cells according to previous study [33], and the details could be found in Supporting Information. We then employed our previously established combined strategy for cell lysis, RNA digestion and nucleosides extraction in one-tube [34]. After incubation at 90℃ for 10 min to allow the lysis of cell, the enzymatic digestion was carried out with the addition of phosphodiesterase I (0.0125 U) and alkaline phosphatase (0.75 U), followed by CMCT derivatization, DSPE with CeO2, and subsequent LC-MS/MS analysis. With this developed method, we can achieve the detection of inosine in a few cells as well as in a single cell (Fig. 4B). The measured content of inosine was 0.26% in a single cell, which is comparable to the results obtained by using RNA samples (0.28%, Fig. 3).
|
Download:
|
| Fig. 4. Determination of inosine in RNA from a single cell and from sleep-deprived mice. (A) Schematic illustration of the analytical procedure for detection of inosine in RNA from a single cell. (B) The extracted-ion chromatograms for the detection of inosine in RNA from a single cell, 2 cells and 4 cells. (C) Quantification of inosine in total RNA of different tissues of sleep-deprived mice (n = 3) and control mice (n = 3). Unpaired t-test was performed for the statistics analysis. Data were presented as means ± SD from three independent experiments. **P < 0.01. | |
Many people are suffering from insomnia that could affect human neural system [35,36]. Long-term insomnia could also cause a variety of other diseases, such as diabetes and heart diseases [37]. We previously found the significantly increased level of inosine at position Chr1:63117284 of Ino80dos RNA of tissues from sleep-deprived mice compared to the control mice [38]. However, the correlation of the overall level of inosine in RNA and insomnia is still unknown. Here, we further applied the developed method to examine the levels of inosine in RNA of tissues (heart, liver and spleen) of sleep-deprived mice as well as in control mice. The results showed that the levels of inosine in all the tested tissues (heart, liver or spleen) were significantly increased in sleep-deprived mice compared to the control mice (Fig. 4C), indicating that inosine is associated with sleep deprivation and might be a potential indicator of sleep disorder.
In summary, we developed a highly sensitive method by combining CMCT derivatization with LC-MS/MS analysis for the detection of inosine in RNA of mammalian cells and tissues. We were able to detect endogenous inosine in RNA as low as 13.0 pg. In addition, inosine in RNA from a single cell could also be distinctly detected due to the improved detection sensitivity after CMCT derivatization. Furthermore, we found that the content of inosine in RNA of sleep-deprived mice was significantly increased compared with the control mice, indicating that inosine may be involved in sleep disorder. Taken together, the CMCT derivatization in combination with mass spectrometry analysis showed great potential in the determination of endogenous modifications from trace amount of RNA as well as from a single cell, which may facilitate uncovering the functions of RNA modifications in the scenario where only limited samples are available.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentsThe work is supported by the National Key R & D Program of China (Nos. 2022YFA0806600, 2022YFC3400700) and the National Natural Science Foundation of China (Nos. 22277093, 22074110, 21721005).
Supplementary materialsSupplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cclet.2023.108243.
| [1] |
I.A. Roundtree, M.E. Evans, T. Pan, C. He, Cell 169 (2017) 1187-1200. DOI:10.1016/j.cell.2017.05.045 |
| [2] |
T. Liu, C.J. Ma, B.F. Yuan, Y.Q. Feng, Sci. China Chem. 61 (2018) 381-392. DOI:10.1007/s11426-017-9186-y |
| [3] |
J. Song, C. Yi, ACS Chem. Biol. 12 (2017) 316-325. DOI:10.1021/acschembio.6b00960 |
| [4] |
M. Frye, B.T. Harada, M. Behm, C. He, Science 361 (2018) 1346-1349. DOI:10.1126/science.aau1646 |
| [5] |
Q. Wang, J.H. Ding, J. Xiong, et al., Chin. Chem. Lett. 32 (2021) 3426-3430. DOI:10.1016/j.cclet.2021.05.020 |
| [6] |
P. Boccaletto, F. Stefaniak, A. Ray, et al., Nucleic Acids Res. 50 (2022) D231-D235. DOI:10.1093/nar/gkab1083 |
| [7] |
X.J. You, S. Zhang, J.J. Chen, et al., Nucleic Acids Res. 50 (2022) 9858-9872. DOI:10.1093/nar/gkac770 |
| [8] |
M.Y. Cheng, X.J. You, J.H. Ding, et al., Chem. Sci. 12 (2021) 8149-8156. DOI:10.1039/d1sc01972d |
| [9] |
M.Y. Chen, Z. Gui, K.K. Chen, et al., Chin. Chem. Lett. 33 (2022) 2086-2090. DOI:10.1016/j.cclet.2021.08.094 |
| [10] |
A.A. Schaffer, E. Kopel, A. Hendel, et al., Nucleic Acids Res. 48 (2020) 5849-5858. DOI:10.1093/nar/gkaa305 |
| [11] |
X.J. You, L. Li, T.T. Ji, et al., Chin. Chem. Lett. 34 (2023) 107181. DOI:10.1016/j.cclet.2022.01.074 |
| [12] |
M.W. Gray, Biochemistry 51 (2012) 5235-5242. DOI:10.1021/bi300419r |
| [13] |
B.L. Bass, Annu. Rev. Biochem. 71 (2002) 817-846. DOI:10.1146/annurev.biochem.71.110601.135501 |
| [14] |
R.W. Holley, G.A. Everett, J.T. Madison, A. Zamir, J. Biol. Chem. 240 (1965) 2122-2128. DOI:10.1016/S0021-9258(18)97435-1 |
| [15] |
K. Nishikura, Annu. Rev. Biochem. 79 (2010) 321-349. DOI:10.1146/annurev-biochem-060208-105251 |
| [16] |
I. Alseth, B. Dalhus, M. Bjoras, Curr. Opin. Genet. Dev. 26 (2014) 116-123. DOI:10.1016/j.gde.2014.07.008 |
| [17] |
Y. Yang, S. Okada, M. Sakurai, RNA Biol. 18 (2021) 999-1013. DOI:10.1080/15476286.2020.1867797 |
| [18] |
X. Xu, Y. Wang, H. Liang, Curr. Opin. Genet. Dev. 48 (2018) 51-56. DOI:10.1016/j.gde.2017.10.009 |
| [19] |
T. Nakahama, Y. Kawahara, Cell. Mol. Life Sci. 77 (2020) 2931-2948. DOI:10.1007/s00018-020-03466-2 |
| [20] |
Y. Dai, B.F. Yuan, Y.Q. Feng, RSC Chem. Biol. 2 (2021) 1096-1114. DOI:10.1039/d1cb00022e |
| [21] |
B. Chen, B.F. Yuan, Y.Q. Feng, Anal. Chem. 91 (2019) 743-756. DOI:10.1021/acs.analchem.8b04078 |
| [22] |
M.C. Owens, C. Zhang, K.F. Liu, Mol. Cell. 81 (2021) 4116-4136. DOI:10.1016/j.molcel.2021.07.036 |
| [23] |
M.Y. Chen, C.B. Qi, X.M. Tang, et al., Chin. Chem. Lett. 33 (2022) 3772-3776. DOI:10.1016/j.cclet.2021.12.008 |
| [24] |
Y.J. Feng, X.J. You, J.H. Ding, et al., Anal. Chem. 94 (2022) 4747-4755. DOI:10.1021/acs.analchem.1c05292 |
| [25] |
R. Zhang, W. Lai, H. Wang, Anal. Chem. 93 (2021) 15567-15572. DOI:10.1021/acs.analchem.1c04151 |
| [26] |
H.Y. Pan, Y. Yu, T. Cao, et al., Anal. Chem. 93 (2021) 14907-14911. DOI:10.1021/acs.analchem.1c03869 |
| [27] |
X.M. Tang, T.T. Ye, X.J. You, et al., Chin. Chem. Lett. 34 (2023) 107531. DOI:10.1016/j.cclet.2022.05.045 |
| [28] |
B.L. Qi, P. Liu, Q.Y. Wang, et al., TrAC Trend. Anal. Chem. 59 (2014) 121-132. DOI:10.1016/j.trac.2014.03.013 |
| [29] |
Q.F. Zhang, H.M. Xiao, J.T. Zhan, B.F. Yuan, Y.Q. Feng, Chin. Chem. Lett. 33 (2022) 4746-4749. DOI:10.1016/j.cclet.2022.01.004 |
| [30] |
J.M. Chu, C.B. Qi, Y.Q. Huang, et al., Anal. Chem. 87 (2015) 7364-7372. DOI:10.1021/acs.analchem.5b01614 |
| [31] |
H. Shintaku, H. Nishikii, L.A. Marshall, H. Kotera, J.G. Santiago, Anal. Chem. 86 (2014) 1953-1957. DOI:10.1021/ac4040218 |
| [32] |
J. Wen, L.A. Legendre, J.M. Bienvenue, J.P. Landers, Anal. Chem. 80 (2008) 6472-6479. DOI:10.1021/ac8014998 |
| [33] |
F.L. Liu, T.T. Ye, J.H. Ding, et al., Anal. Chem. 93 (2021) 6848-6856. DOI:10.1021/acs.analchem.1c00915 |
| [34] |
W. Huang, C.B. Qi, S.W. Lv, et al., Anal. Chem. 88 (2016) 1378-1384. DOI:10.1021/acs.analchem.5b03962 |
| [35] |
T.M. Prince, M. Wimmer, J. Choi, et al., Neurobiol. Learn. Mem. 109 (2014) 122-130. DOI:10.1016/j.nlm.2013.11.021 |
| [36] |
J.E. Kang, M.M. Lim, R.J. Bateman, et al., Science 326 (2009) 1005-1007. DOI:10.1126/science.1180962 |
| [37] |
M. Guidolin, M. Gradisar, Sleep Med. 13 (2012) 779-786. DOI:10.1016/j.sleep.2012.03.016 |
| [38] |
J.H. Ding, M.Y. Chen, N.B. Xie, et al., Biosens. Bioelectron. 219 (2023) 114821. DOI:10.1016/j.bios.2022.114821 |
 2023, Vol. 34
2023, Vol. 34 

