b Institute of Agro-product Safety and Nutrition, Zhejiang Academy of Agricultural Sciences, Hangzhou 310021, China;
c State Key Laboratory of Coordination Chemistry, Nanjing University, Nanjing 210023, China
The mitochondrion is one of the most well studied organelles. It is a double-membrane-bound organelle that is possessed in almost all eukaryotic cells and consist of four parts, namely the mitochondrial outer membrane, inner membrane, interstitial membrane, and mitochondrial matrix. The inner membrane of mitochondria folds inwards to form mitochondrial crest, which is responsible for many biochemical reactions [1]. Mitochondria are small but extremely powerful, which is the main production site of oxidative phosphorylation and synthesis of adenosine triphosphate (ATP) in cells, providing energy for cell life activities, known as the factory of capacity. Mitochondrial activity is essential for all mammalian tissues and for most cell types (excluding red blood cells) [1, 2]. In addition to supplying energy to cells, mitochondria are involved in processes such as cell differentiation, cell messaging and apoptosis, and have the ability to regulate cell growth and cell cycle [3]. Mitochondria also play a central role in intermediate metabolism, synthesis of key iron-sulfur cluster enzymes, cellular calcium homeostasis, and cell death through apoptosis. In addition to water, mitochondria are composed of nucleic acids, proteins and lipids, which play an important role in cells. The disorder of genetic factors controlling mitochondrial gene replication and transcription can lead to the generation of disease factors or even death of the body [4, 5]. If the mitochondrial electron transport chain does not capture electrons correctly, the escaped electrons will combine with oxygen molecules to generate superoxide free radicals, which can easily destroy the mitochondrial DNA (mtDNA) bases and cause mtDNA mutations, and then accumulate some aging or disease factors of cells, such as diabetes, heart disease, arthritis [6-9]. In addition, these superoxide radicals activate inflammation, causing damage to other tissues in the body [10-15]. Differences between the mitochondria in normal cells and cancer cells include a number of functional changes, such as mtDNA mutations that cause oxidative phosphorylation inhibition, nutrient hypoxia or depletion. Specially, changes in mitochondrial metabolism in cancer cells are conducive for selective targeted treatment of mitochondria. These treatments are mainly aimed at the specific characteristics of the mitochondria found in cancer cells. Therapeutic drugs that specifically target mitochondria are an effective treatment method for diseases. For this reason, engineered therapeutics targeting the mitochondria are viewed as potential disease treatment strategies.
Therefore, mitochondria play an important role in cell growth and development. Differences in mitochondria between normal and abnormal cells include many functional changes. The change of mitochondrial metabolism is conducive to selective targeted therapy of mitochondria. In recent years, many therapeutic drugs and targeting systems for mitochondrial diseases have been developed, making important contributions to clinical research on the diagnosis and treatment of mitochondrial diseases. Herein, we provide a comprehensive review of recent advances in the rational design of mitochondrial targeting nanocarriers and discuss the different strategies available for mitochondrial targeting nanocarriers. We also highlight the advantages and disadvantages of the various carrier systems currently in use. Finally, the application prospect of a new generation of mitochondrial targeted drug delivery system in the field of drug therapy is prospected. We hope that this review will offer a comprehensive understanding of the importance of mitochondrial targeting drug delivery systems and facilitate the development of more effective mitochondrial targeting systems.
2. Current therapeutic drugs for mitochondrial diseasesCurrent therapeutic drugs for mitochondrial diseases include active proteins, genes, and small-molecule drugs, etc. Protein- and peptide-based drugs usually have specific three-dimensional structures and sites of action, which can exert specific therapeutic effects in the body. Proteins are suitable for use as a sustainable controlled release formulation [16]. The advantages of sustained-release protein delivery formulations include its suitability for long-term treatment, use for local delivery, fewer side effects, less doses than traditional drugs, reduced dosing frequency, and improved patient compliance. They have better clinical effectiveness and safety compared to traditional drugs. However, when these drugs enter the body, most of them have problems such as poor stability, poor circulation, low bioavailability, short half-life in vivo, and easy to be degraded by enzymes in the body. In particular, protein and peptide drugs that are employed to treat chronic diseases often require long-term and frequent injections, which not only result in low treatment efficiency, but may cause other adverse reactions. Therefore, the pharmaceutical industry continues to carry out research to improve the performance of protein and peptide drugs, such as the modification of hydrophilic polymers on proteins to enhance the water solubility of protein drugs, which provides the basis for the application of most protein drugs in vivo. The modified protein drug has increased molecular weight and avoids glomerular filtration. Meanwhile, the hydrophilic polymer blocks reticulo-eneophagocytic system (RES), which exists widely in liver, spleen and lymph nodes, and provides a good protection for the protein drug. The protective effect of hydrophilic polymers also prevents the degradation of enzymes in the body. What is more, the chemical bonds between the drug and the polymer slowly hydrolyze over time, which facilitates the release of the protein drug [16-18]. The ideal protein delivery formulation should have the function of controlled release, maintain the therapeutic level and ensure the stability of the encapsulated protein during the prolonged drug action time. The components of the delivery system must be biodegradable and biocompatible, non-immunogenic, and non-toxic. At the same time, the structure and activity of the protein drug must not be changed during the preparation of the delivery system, and the protein must assume an active conformation after being released in the body. On top of all these factors, the preparation method for protein intracellular delivery should have scalability, durability and reproducibility. In fact, the use of carriers to deliver encapsulated proteins to target organelles such as mitochondria is one of the current research hotspots.
Another therapeutic for mitochondrial diseases is gene-based drugs. Gene-based therapies can effectively regulate both coding and non-coding target genes with high specificity, and have broad application prospects in the treatment of various human diseases [19-21]. Biotech and pharmaceutical companies are working hard to develop many gene therapy drugs. Gene therapy drugs are also biomedical products that are administered in the form of nucleic acids, viruses, genetically engineered microorganisms, or lipid complexes to achieve preventive, diagnostic and therapeutic effects [22]. The approach of gene therapy represents the latest advance in the revolution in biomedical technology. In the past half century, gene therapy has rapidly absorbed almost all the advanced biotechnology, and many promising human gene therapy drugs have been developed. Since the end of the last century, a variety of gene therapy drugs, including cell-mediated gene therapy products, viral vectors, non-viral vectors and naked nucleic acids, have been approved by the US Food and Drug Administration (FDA) for the treatment of human diseases [22]. One current approach for gene therapy involves cloning DNA sequences that encode genes associated with mitochondrial-targeting signals. The resulting recombinant genes are expressed as proteins once they are delivered to the cell. Thereafter, the protein is directed to mitochondrion by the mitochondrial targeting signal (MTS). The MTS enters cells through mitochondrial import mechanisms [23-27]. The advantages of combining gene therapy vectors with encoded MTS include not only the ease of cloning of recombinant DNA, but also the advantage that these constructs can be prepared without chemical modifications. Although the prospect of gene drugs is promising, there are still many problems should be solved, such as safety, transmissibility, selectivity, stability, immunity. Therefore, the development of the nucleic acid drugs and their safe and efficient delivery system are the key challenges in the clinical application of gene therapy.
In the last few years, several new candidate drugs have been assessed and developed for various diseases. Among them, the structure of small-molecule drugs has good spatial dispersion, and its chemical properties determine its good drug-making properties and pharmacokinetic properties. These characteristics make small molecule drugs show great advantages in the drug development process and other drug fields, and the research and development of small molecule drugs is increasingly favored by the market.
Meanwhile, small-molecule compounds derived from natural products have been demonstrated to display good preventive and treatment effects due to their antioxidant, anti-inflammatory, and neuroprotective functions [28, 29]. However, small molecule drugs, especially anti-tumor drugs, often have problems such as off target, low solubility, high toxicity, rapid excretion or unstable biological distribution. At the same time, most natural small molecule drugs have inherent disadvantages such as short half-life, poor stability in vivo, easy to be cleared, which make it difficult for small molecule drugs to accumulate effectively at the target site [28-30]. At present, researchers use nanoparticles as drug delivery carriers, which can effectively improve the water solubility of small molecule drugs, thereby reducing the drug dose and cytotoxicity. The drug can be delivered to the lesion to improve the efficacy. It also plays an important role in reducing clearance and increasing drug retention time in the body [31].
Actually, by targeting mitochondria delivery, some of disadvantages for the therapeutic drugs have been solved recently. Until now, their application in various targeted therapies, such as targeted therapies for cancer, neurodegenerative diseases, autoimmune diseases, and several other diseases, has been reported [32, 33]. The development of mitochondria-targeted drug therapy in the pharmaceutical industry is growing. In subsequent sections, we introduce of mitochondria-targeted moieties and present delivery strategies for achieving targeted delivery to the mitochondria, with focusing on rationally designed routes, related mechanisms, and the potential therapeutics.
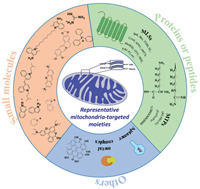
3. Mitochondria-targeted moietiesSince mitochondria are considered as emerging pharmacological targets, there are currently several targeted therapeutic strategies for mitochondria-related diseases. Based on the characteristics of the mitochondrial membrane, numerous of mitochondria-targeted moieties have been developed for enhancing the mitochondrial uptake (Fig. 1) [34-55].
|
Download:
|
| Fig. 1. The three kinds of mitochondria-targeted moieties designed for enhancing the mitochondrial uptake. | |
Drugs that regulate mitochondrial function to achieve certain diagnostic or therapeutic effects can be used for mitochondrial targeting, but a series of problems such as the feasibility of drug delivery, drug toxicity and biocompatibility need to be considered. In order to solve the above problem, designing a series of mitochondrial targeted nanocarriers based on small molecule moieties is developed by using the high negative potential of the properties in the mitochondria membrane. The negative potential of plasma membrane size is only 30–40 mV, while mitochondrial membrane negative potential can reach 120–180 mV, significantly less than the plasma membrane potential, making the Coulomb force between the positively charged compounds and mitochondrial membrane greater than that of other negatively charged membrane. They are more likely to accumulate in the mitochondria. For example, the enrichment amount or concentration of triphenylphosphine (TPP) cationic compounds in mitochondria is 20-300 times higher than that in plasma membrane, achieving highly specific targeting of mitochondria [7, 34]. However, some lipophilic cations have phototoxicity and poor water solubility, leading to poor circulation in vivo and reducing their efficacy. The mitochondrial targeting section of the widely studied TPP ion derivative takes advantage of its strong lipophilicity and the properties of delocalized cationic to target inner mitochondrial membrane (IMM). The negative membrane potential of IMM is the key to driving lipophilic cations to target mitochondria. Therefore, in addition to TPP cation derivatives, aromatic heterocyclic cation derivatives such as pyridine cation and quinoline also have mitochondrial targeting ability [47, 53]. Dequalinium (DQA) is a small positively charged molecule containing two cationic amino quinoline rings, which are attracted to the IMM through electrostatic interaction and selectively accumulate in the mitochondria-cartilage matrix [47]. Under physiological conditions, guanidine or bisguanidine are strong bases (pKa ~13) with a positive charge delocalization. When guanidine or bisguanidine groups are partially coupled to the target pharmacokinetic molecule as mitochondrial targets, a delocalized positive charge is introduced to increase lipophilicity, thus concentrating the target molecule in the IMM [48, 49]. Rhodamine contains three benzene rings in the form of spironolactone in a nonpolar solvent [50]. In a polar solution, it changes from the lactone form to a zwitter ion and exists in the form of ammonium cation in the ring-open state. Due to the negative potential of IMM, rhodamine is able to specifically target mitochondria in the form of ammonium cations. Tetramethylrhodamine-5-isothiocyanate (TRITC) is another hydrophilic derivative of rhodamine which is often used as a fluorescent probe to target mitochondria [51, 52]. Based on the lipophilic and cationic properties, pyridine salts structures show the ability to target mitochondria [53]. Berberine (BBR), an isoquinoline alkaloid, has many outstanding pharmacological effects. Because anthocyanin dyes are cationic and lipophilic, they can accumulate in the mitochondria as well [40, 41]. Herein, the currently reported small molecule moieties are summarized in Table S1 (Supporting information). According to the properties of mitochondrial membrane, we believe more and more small molecules moieties will be designed for mitochondrial targeting.
3.2. Peptide/protein moietiesAs summarized in Table S2 (Supporting information), another strategy for targeting mitochondrial involves using peptides/proteins moieties, which have specific physical and chemical properties. Similar to lipophile cations, mitochondria-targeted peptides (MTP) is an effective alternative to lipophile cations and can target bioactive substances to the mitochondria. The synthetic MTP contains hydrophobic amino acids (tyrosine, phenylalanine, and isoleucine) which can penetrate the lipid membrane and positively charged amino acids (lysine and arginine) by binding to the negatively charged mitochondrial membrane through electrostatic interactions [53-55]. Currently, most synthetic MTP structures contain positively charged amino acids (arginine and lysine) in order to concentrate on the negatively charged IMMs. The MTPs are firstly recognized by the outer mitochondrial membrane (OMM) once they attach to the pharmacophore. They then enter the mitochondrial matrix via OMM, thereby achieving mitochondrial targeting capabilities. At present, the most commonly used MTP mainly include pro-apoptotic peptide (KLAKLAK)2, biologically active peptide sequence (rFxrFxrFxr), and the trans-activating transcriptional activator (TAT). (KLAKLAK)2 is a kind of cationic amphiphilic peptides which have α-helix configuration [53]. It has been reported that KLA is a pro-apoptotic peptide, which can enrich mitochondria, destroy the negatively charged mitochondrial membrane and induce cell apoptosis. The bioactive peptide sequence rFxrFxrFxr possesses d-arginine and mitochondrial targeting ability [56]. The arginine-rich cell-penetrating peptide, TAT (YGRKKRRQRRR) acts as a mitochondrial targeting element that targets molecules and localizes in mitochondria [57-63]. Another mitochondrial penetrating peptide, namely mitochondrial localization sequences (MLSs), usually consists of 20–30 amino acids (detailed sequences in Table S2) [64-81]. MLSs can be recognized by mitochondrial surface receptors, and the targeting sequences are variable in length and amino acid sequence. Their main characteristics are the presence of positively charged residues, the lack of acidic residues, and the formation of amphiphilic secondary structure. Mitochondrial targeted receptor proteins form a protein family, including the translocases of outer membrane (TOM) and the translocases of inner membrane (TIM) [53, 63]. Protein targeting sequence has good biocompatibility, but false positive may occur, which may target to other locations other than mitochondria, resulting in false positive signals. The protein sequence is eliminated too quickly in the body to be effective. To solve this problem, some positively charged amino acids can be linked to targeted protein sequences to increase the efficiency of mitochondrial targeting.
3.3. Other moietiesAmphiphilic modifications are often carried out on nanoparticles to make it easier for lipophilic substances to pass through the lipid components of mitochondrial membrane. Amphiphilic polymers provide chemically reactive groups for post-modification of polymer nanomaterials with targeting ligands (such as antibodies and peptides) to specifically target cell sites (such as membrane receptors and organelles) [82]. Chitosan and chitosan-based nanoparticles, which are examples of polymers, have a number of unique properties, including low toxicity, biocompatibility, biodegradability, and the capacity for mucoadhesion. Chitosan is a positively-charged polycationic biological macromolecule that can be developed as mitochondrial drug delivery systems for mitochondria [9, 83, 84].
In addition to polymers moieties, there are other types of targeted modification groups, such as aptamers, transition metal complexes, which are summarized in Table S3 (Supporting information). Aptamer, which is a single-stranded DNA or RNA (ssDNA or ssRNA), has a unique tertiary structure that can specifically bind to homologous molecular targets. Aptamers are usually screened through systematic evolution of ligands by exponential enrichment [85, 86], targeting from small molecules [87, 88] to biomacromolecules [86, 89-94], infected cells, stem cells, and tumor tissues [95-104]. Aptamers can be used as therapeutics by themselves. They have been extensively studied as targeting ligands such as for mitochondria. The programmability of aptamers and the ease of synthesis and modification allow them to be universally used as targeted ligands for drug delivery [105-108]. Previous studies have reported that many Ir(Ⅲ) complexes have a positive charge and can specifically target mitochondria without modification of the mitochondrial-targeting units [39]. In addition, some natural substances or designed molecular substances can interact with the mitochondrial respiratory chain complex to generate a large number of reactive oxygen species (ROS) and oxidized endogenous pyridine nucleotides, triggering the opening of mitochondrial permeability pores (MPTP). Glycyrrhizic acid (GA), an extract from licorice, is a mitochondrial targeting ligand with a near-neutral charge [28]. GA can interact with the mitochondrial respiratory chain complex to produce large amounts of hydrogen peroxide and oxidize endogenous pyridine nucleotides, triggering MPTP to open. The drug then moves inside the mitochondria to react. Kong's group found hypericin (HY) is an effective ligand to target mitochondria [29]. Under laser irradiation, HY interacts with mitochondrial respiratory chain to produce singlet oxygen. The generated singlet oxygen then oxidizes critical thiol groups and endogenous pyridine nucleotides, resulting in the opening of mitochondrial permeability transition pores, then the drug goes into the mitochondria, which is a common mechanism of action for various mitochondrial-targeting ligands [109, 110].
4. Nanoparticle-mediated delivery systemsA wide array of nanocarriers have been identified as promising strategies for organelle-targeted delivery. In these strategies, the delivery systems could be engineered with various ligands for targeting to the desired subcellular compartments like cytosol, nucleus and mitochondria [111-118]. Moreover, recent studies have reported that engineered nanocarriers can be combined with integration of stimuli triggers to allow for drug delivery with both dosage-dependent and spatiotemporal-controlled manners possible, facilitating release of drug with desirable pharmacokinetics [119]. Until now, numerous synthetic nanosystems have been designed and developed for mitochondria-targeted drug delivery [120-124].
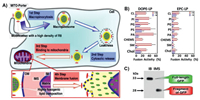
4.1. LiposomeDue to their relative safety and simplicity, lipid-based nanoparticles have been widely used in systemic delivery nanocarriers. The liposome carrier can transport water-soluble drugs in its core or fat-soluble compounds in its outer membrane layer. Compared with chemically-modified drugs, this delivery method encapsulates active molecules without altering their molecular structure, thereby maintaining the pharmacy dynamic characteristics of the drug. This type of delivery system protects the loaded drug from biodegradation or chemical degradation in the blood, and releases drugs at the target site in a suitable manner [125]. In current clinical trials with mice and humans, liposome drug delivery systems have been used for targeted delivery of a variety of anticancer drugs, including biomacromolecules and hydrophobic drugs [62, 125-147]. Some liposomes, such as liposomal adriamycin and liposomal paclitaxel have been approved by FDA and are widely used in the treatment of metastatic ovarian cancer, breast cancer. This delivery system also significantly reduces toxicity of the drugs in the patient's heart [125, 127-139]. To prevent the enrichment of liposomal nanoparticles in the organism, which makes their degradation difficult, their surface generally modified with a little hydrophilic group to improve their biocompatibility. Chemical modifications on liposome can enable their effective entry into mitochondria. For example, Harashima's team described a liposome carrier that delivered a macromolecular cargo to the interior of the mitochondria via membrane fusion [137]. As shown in Fig. 2, these liposomes, called MITO-Porters, were modified with octaarginine (R8) moieties on their surfaces as a complete vesicle MITO-Porter. Amino acid polymers and synthetic arginine peptide analogues have been reported to be as the translocation domains to improve cargo delivery efficiency [140-142]. As shown in Fig. 2A, MITO-Porter entered the cell through endocytosis, was degraded by lysosomes, and banded to mitochondria through electrostatic interaction. The results showed that nanocarriers containing R8 have better fusion efficiency than those without R8, indicating that the strong electrostatic binding between R8-based liposomes and mitochondria stimulates. The delivery model of this experiment was green fluorescent protein, and western blot showed that it had a good delivery effect. This provides guidance for the mitochondria targeted drug delivery of biomacromolecules. The MITO-Porters can be employed to transport functional nucleic acids, proteins, and small biologically active molecules. Such delivery systems are very promising as they can transport molecules with different physicochemical characteristics as well as large or small encapsulated molecules. Cationic liposomes are considered to be another promising nanocarriers for improving mitochondrial delivery. For example, Taillandier's team used trimethyl aminoethane carbamoyl cholesterol iodide (TMAEC-Chol) and dioleoyl phosphatidylethanolamine (DOPE) to create a liposome with clear size and stability [143]. In order to further transfer the oligonucleotide into the mitochondrial matrix, the oligonucleotide was linked to the ornithine transcarbamylase signal peptide, which is recognized by the mitochondrial protein import mechanism and then enters the mitochondria. Therefore, suitable cationic liposomes and peptide oligonucleotide conjugates can be used in mitochondrial gene therapy experiments, which lays a foundation for the treatment of mtDNA abnormal diseases [144, 145]. Deng's group postulated that mitochondrial targeting and stimuli-triggered liposome delivery system could be employed to derive new therapies for deep tumor treatment [135]. Liposomes were connected to TPP with verteporfin and gold nanoparticles being encapsulated inside the liposomes. Low-dose X-rays can trigger verteporfin to produce cytotoxic ROS. In this process, porphyrin transferred the absorbed energy to oxygen to form a highly active single oxygen with a short-term maintenance time. Single oxygen destroyed various biological structures within its diffusion range. This process leaded to local vascular atresia, cell destruction, and cell death under certain circumstances. Co-encapsulation of gold nanoparticles (10 nm) in the central core of liposomes can increase the production of ROS for deep tumor treatment.
|
Download:
|
| Fig. 2. (A) Mechanism of the transport of macromolecules to the mitochondria using MITO-Porter. (B) R8-LPS (a fusion lipid commonly used for drug delivery) composed of DOPE (R8DOP-LPS) shows higher fusion activity than R8-LPS (R8-EPC-LPS) composed of egg yolk phosphatidyl choline. (C) The purified GFP was incubated with the IMS portion or IB portion, and Western blotting showed that the molecular weight of GFP was 28 kDa when incubated in IMS, but remained intact when incubated in IB, thus proving that Mito-Porter delivered GFP to IMS via OM. Reproduced with permission [137]. Copyright 2018, Elsevier. | |
In conclusion, lipid-based nanoparticle delivery systems for mitochondria have been extremely studied owing to their promising features. However, some limitations, such as the high cost of raw material, easy accumulation in the phagocytes are still the main issues for this strategy [146, 147]. Thus, other alternative materials are needed for specific disease, as well as offer unique advantages.
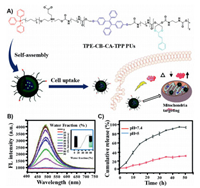
4.2. Polymer nanosystemOwing to their simple preparation method and easy modification, high-molecular-weight, diverse polymeric delivery systems have been extensively used for drug delivery and achieved excellent performance. Especially, through modifying with mitochondria targeted moieties, the polymer-based nanocarriers such as polyethylene glycol (PEG), polycaprolactone (PCL), and chitosan have been designed for the treatment of mitochondria related disease [9, 67, 148-181]. For example, Du's group reported an innovative mitochondria targeted prodrug nanocarrier characterized by pH-triggered and aggregation-induced emission (AIE) properties (Fig. 3) [154]. In this work, the nanocarrier was constructed by surface-modification of the TPP group with polyurethane (PU). Co-localization studies of mitochondrial pairs showed that this modification can improve the targeting of organelles. A zwitterionic shell was then attached, which can prolong the blood circulation and accumulate at the tumor site. The ketal bond of the nanomicelles enabled controlled release of therapeutically significant molecules. In addition, the addition of luminogen tetraphenylethene (TPE) based on AIE was allowed to realize labelling and monitoring of the mitochondria of living cells in real time. The model drug cinnamaldehyde induced ROS production in tumor cells to promote oxidative stress and activate apoptosis pathways. Moreover, this nanocarrier has been proven to have many advantages in cell imaging, including good AIE properties, anti-photo-bleaching and low cytotoxicity, which provide a feasible strategy for simultaneous cancer diagnosis and treatment. In summary, this prodrug nanocarrier not only has mitochondrial targeting function but also can kill cancer cells and monitor the process of apoptosis in real time. Meantime, Chen's team created a polymer-based nanoparticle based on TPP-poly(ethylene glycol)-poly(ε-caprolactone) (TPP-PEG-PCL) polymers that enhanced mitochondrial targeting with TPP [155]. Gambogic acid was used as model drug to treat lung cancer. Due to poor water solubility (~10 µg/mL) and short half-life (15.7 min), the low bioavailability of gambogic acid limits its clinical application [9]. The limitation of gambogic acid was overcome when the polymer nanomicelles were formed. After the gambogic acid was transferred into mitochondria, it can induce apoptosis of cancer cells by inhibiting human Bcl-2 family proteins and activating cystease. Recently, Choi's team created an amphiphilic polymer nanomicelle using glycol-chitosan and DQA [9]. In this nanosystem, ethylene glycol chitosan (GC) was used to enhance in vivo circulation and DQA served as mitochondrial targeting and wrap around the anticancer drug curcumin for cancer treatment studies. The molecular structure of curcumin has phenolic hydroxyl groups, which can capture or clear free radicals and have antioxidant effects [10]. Zhang et al. found that curcumin could inhibit the expression of COX-2 mRNA and prevent the occurrence of tumor [11]. The defects of fast metabolism and short half-life of curcumin were ameliorated by targeting mitochondria delivery. The experiment showed that this nanosystem had the great potential of mitochondrial targeted drug delivery for cancer treatment.
|
Download:
|
| Fig. 3. (A) Mechanism of the pH-triggered prodrug. (B) In order to observe the AIE properties of nanocarriers, the fluorescence changes of prodrug polymer in DMSO/water system were studied. The experiment showed that the fluorescence gradually increased with the increase of water content, indicating the desire to be aggregates. (C) The results of experiment results proved that the pH-responsive drugs of TPE-CB-CA-TPP were beneficial to the effective release of intracellular drugs. Reproduced with permission [154]. Copyright 2019, The Royal Society of Chemistry. | |
Due to the good biodegradability and biocompatibility, numerous polymers, specially the synthetic polymers can be referred as biomaterials to produce nanoformulations for mitochondria-targeted delivery. However, owing to the complexity of polymer-based nanocarriers with multicomponent 3D structures, careful designing and construction are needed to make sure reproducible formulations in the large-scale manufacturing processes [79, 181]. Furthermore, the smart polymers with multifunction, multicomponent and controlled release on demand in mitochondria and high efficient diffusion of therapeutic drugs through a stimulus property of the disease microenvironment are urgently needed.
4.3. Inorganic nanosystemWith the development of nanotechnology in recent years, inorganic nanocarriers with unique biological, chemical, and physical properties have become increasingly prominent in biomedicine. Inorganic nanocarriers are utilized not only as diagnostic reagents, but also for the prevention and treatment of certain medical conditions such as tumor, cardiovascular diseases and neurodegenerative diseases. Presently, an increasing number of inorganic nano-medicine carriers have been employed as targeted therapies for specific tumors. Accordingly, they have gradually become a research focus in the medical field [182-203].
Due to the biocompatibility and tunable pore size, mesoporous silica nanoparticles (MSNs) have been applied for storage and delivery of drugs [199]. Jung's team firstly conjugated guanidinium derivative to MSN shell with Fe3O4 core and then coated it with the anti-tumor drug doxorubicin (DOX) to achieve mitochondrial targeted tumor therapy [182]. DOX blocks DNA synthesis by inserting DNA strands, and its toxicity to topoisomerase also inhibits DNA synthesis. DNA damage leads to activation of p53 tumor suppressor gene, aggregation of related proteins, and mitochondrial dysfunction, which are a series of events in the early stage of cell death. DOX also induces the production of free radicals and accelerates the induction of tumor cell death. Jung's experimental results showed that the nanosystem could localize in mitochondria and release DOX on demand, which accelerated the induction of tumor cell death.
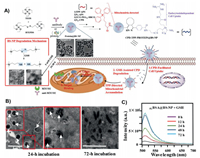
Current methods for mitochondria targeting are feasible for small-molecule drugs, while many methods are not suitable for large drugs, such as proteins (including antibodies). The first example of using biodegradable silica nanoparticles (BS-NPs) to target mitochondria for intracellular delivery of native proteins (and antibodies) was reported by Yao's team (Fig. 4) [184]. In this work, the synthesized biosilica nanoparticles were modified with cell-penetrating poly(disulfide) (CPD) that allowed nanoparticles to enter cells, and TPP drives for targeting mitochondria. In the presence of glutathione (GSH), CPD was degraded and TPP was exposed, and then the BS-NPs could enter into the mitochondria for the protein releasing. The experimental results provided reference for the delivery of macromolecules into mitochondria.
|
Download:
|
| Fig. 4. (A) Preparation and application of cell-permeable CPD-TPP-protein@BS-NP. (B) HeLa cells were incubated with CPD-TPP-RBBSA@BS-NP. The results showed that after cultured for 24 h, most BS-NPs were located in the mitochondria (white circles denote mitochondria, arrows denote BS-NPS), and degraded after 72 h. (C) To monitor the release of BSA on the nanocarrier, the supernatant of GSH-treated FLBSA@BS-NPs was collected periodically between 0 and 72 h for fluorescence measurements. The experiment showed that the nanocarrier encapsulated BSA was successfully released. Reproduced with permission [184]. Copyright 2019, Wiley. | |
Various inorganic nanoparticle-mediated delivery systems have been studied for mitochondria targeted delivery of drugs. Unlike the lipid-based nanoparticles, these inorganic nanoparticles are considered to be easier during manufacturing. The stability of the inorganic nanoparticles enables them to remain in long-term circulation in the body, however, this also makes them difficulty to be decomposed and cleared from body, leading to the accumulation in the liver, lungs and kidney [200]. Therefore, development of the biodegradable inorganic nanoparticle-based system is an effective way to solve this issue.
4.4. Peptide/protein systemDue to peptide/protein-based NPs have the advantages of high bioavailability, good solubility, low toxicity, and good targeting ability, they have been developed as one of the most promising nanomaterials. In addition, the peptide-based backbone exhibits the following advantages: easy synthesis, high tunability, good biocompatibility, and high absorbability in cells or in vivo. Due to their simple and modular synthesis, peptides that target the mitochondria can adapt to different delivery conditions. The main design features of peptides targeting the mitochondria are cationic charge and lipophilicity. Cationic charge enables the peptides to fully utilize the negative mitochondrial membrane potential while lipophilicity enables them to interact well with the hydrophobic membrane of the mitochondria. As carriers, peptides are covalently linked to therapeutic drugs to enhance their drug value and unravel a pathway for the study of mitochondrial biology [31, 57, 62, 151, 194, 204-215].
Recently, Rapaport's team synthesized a polypeptide-peptide nanoparticles (Pop-NPs) with an amphiphilic and cationic β-sheet peptide (PFK) drive [133, 205]. After complexed with siRNAs via charge interaction, the nanosystem could deliver oligonucleotide cargos to mitochondria, which provided guidance for the delivery of nucleic acid drugs.
Protein-based delivery platforms show exciting ways of significantly improving therapeutic effect and reducing adverse reactions in the body [214]. Until now, a number of proteins have been developed for use substrates in protein-based nanoparticles for drugs delivery, such as gelatin, albumin, collagen, whey proteins, casein, gliadin, elastin, silk proteins, soy proteins, and lectins [215]. Lin's team discovered a new self-assembled metalloprotein nanoparticle, GST-MT-3 (Fig. 5). Here, metallothionein-3 (MT-3) can regulate zinc ion homeostasis and control cell proliferation and differentiation by influencing mRNA production [194]. The nanoparticles were linked to mitochondrial transport peptides GST for mitochondrial targeting. After further chelating cobalt ions [GST-MT-3(Co2+)], this nanosystem could induce ROS production and reduce the mitochondrial membrane potential. In addition, GST-MT-3 covalently bonded to paclitaxel could furhter inhibit tumor growth. The in cell and in vivo results showed that the nanoparticles could rapidly assemble in tumors and target mitochondria, inducing ROS production and tumor cell death through the apoptotic pathway.
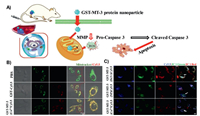
|
Download:
|
| Fig. 5. (A) Illustration mechanism of the novel GST-MT-3 protein nanoparticle. (B) GST-MT-3(Co2+) was rapidly internalized by HeLa and U87MG cells and localized in mitochondria. (C) The mitochondrial membrane potential of HeLa cells decreased after treatment with GST-MT-3-CY5.5 analogues for 24 h. Reproduced with permission [194]. Copyright 2019, Wiley. | |
Protein-based delivery platforms for targeting mitochondria display exciting ways to significantly improve drug delivery and therapeutic effects. The efficacy of protein-based nanoparticles can be increased by combining these nanoparticles with other FDA-approved treatments. While the progress is still needed to develop the potential in the improvement and mechanistic understanding, as well as the feasibility of using protein-based nanoparticles and their derivative in combination therapy in the clinical field of tumor treatment in vivo.
4.5. Other delivery systemsApart from the above nanocarriers employed to transport biologically active molecules into the mitochondria, there is increasing interest in the use of other systems in the delivery of various drugs into mitochondria [34, 46, 105, 133, 146, 163, 197, 216-280]. For example, metal-organic frameworks (MOFs) in particular have arisen in the last years as favorable candidates for nanomedicine applications, owing to their unique properties [217-223]. To date, a number of therapeutic drugs have been encapsulated in MOFs, including anticancer, antibacterial, and antiviral drugs, as well as nucleic acids and biological gases [224]. Recently, Jimenez's group reported a novel mitochondria-targeted MOF-based nanosystem that greatly increased the efficacy of a model cancer drug [234]. A Zr-MOF loaded with the cancer drug dichloroacetate (DCA) and conjugated with TPP for localizing to mitochondria was constructed. Whole transcriptome analysis of cells indicated wide spread changes in gene expression and profound effect on cell physiology and that are related to cell death. They showed that targeting MOFs toward mitochondria represents a valuable strategy for the development of new drug delivery systems.
Different from the introduction of mitochondria targeted moieties, Lin's group has synthesized a nano-MOF for mitochondria-targeted radiation therapy for tumors [235]. In this work, the MOFs with surface modification of cationic ruthenium (Ru) can directly target mitochondria. In this way, targeting delivery with photosensitizers can effectively produce ROS, depolarize mitochondrial membrane potential, and lead to programmed cell death of tumor cells (Fig. 6).
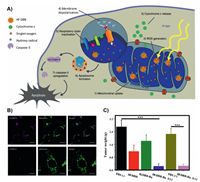
|
Download:
|
| Fig. 6. (A) Process schematic of the Hf-DBB-Ru. (B) Representative mitochondria co-localization images of Hf-DBA-R and Hf-DBB-Ru by CLSM. (C) In vivo anti-cancer efficacy of mitochondria-targeted RDT. Excised tumour weights on day 22. Reproduced with permission [235]. Copyright 2018, Springer Nature. | |
In addition, Kubi et al. also presented a class of amphiphilic cell-penetrating motifs (CPMs). Each of them was composed of four guanidine groups linked to one or two aromatic hydrophobic groups (naphthalene), which was used for targeting mitochondria. They could assemble into nanoparticles via a central scaffold (such as a benzene ring) [229]. CPM transferred drug geldanamycin into cells and allowed them to accumulate in the mitochondrial stroma. The results showed that CPM was metabolically stable and had little toxicity to normal cells, and could be used to deliver therapeutic agents into the mitochondrial matrix to treat mitochondrial diseases.
5. Conclusion and prospectIn summary, various nanocarriers have been developed for mitochondria-target drug delivery and shown great promise for enabling the current and next generations of therapeutics. This review described the current progress of research on nanocarriers for mitochondria-targeted delivery. We herein classify and summarize the reports according to the type of nanocarriers, which is shown in Table S4 (Supporting information). The occurrence and development of many diseases are caused by mitochondrial dysfunction. The subcellular targeting strategy could achieve maximum therapeutic responsive as well as avoiding cytotoxic side effects of the drugs. As a result, drug therapies targeting mitochondria are highly desirable.
To improve drug intracellular delivery and active targeting, especially for the organelle and subcellular drug targeting, the surface of nanocarriers could be incorporated with receptor targeting ligands (e.g., TPP, cationic peptides) to specifically bind with a receptor of the mitochondria membrane. Although multiple basic research studies have been conducted many basic researches on mitochondrial diseases, the design of effective delivery methods need to be further improved. Many challenges currently remain, such as the stability and water solubility issues for liposomes, low drug loading efficacy for the polymeric nanoparticles, and difficulty of being decomposed and cleared from the liver, lungs and kidneys for inorganic nanocarriers, etc.
Different types of nanocarriers have been recognized to possess their own advantages and disadvantages. More mitochondria targeted moieties and interaction mechanisms should be investigated for continuous improvements of novel nanocarriers in high efficiency, high drug loading capacity, strong targeting ability, and low side effects. Future research should also focus on the discovery and design of safer and more efficient mitochondrial-specific nanomaterials.
In conclusion, we believe the potential for drug delivery using mitochondria-targeted nanocarriers is tremendous. More and more novel material or delivery vehicles emerge and others will likely be uncovered through further scientific research.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentsWe gratefully acknowledge financial support from the National Key Research and Development Project (No. 2020YFA0709900), the Key University Science Research Project of Jiangsu Province (No. 19KJA520005), the National Natural Science Foundation of China (No. 22077101), the Open Project Program of State Key Laboratory of Coordination Chemistry (No. 202008).
Supplementary materialsSupplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cclet.2022.02.047.
| [1] |
L. Craven, C.L. Alston, R.W. Taylor, D.M. Turnbull, Annu. Rev. Genom. Hum. Genet. 18 (2017) 257-275. DOI:10.1146/annurev-genom-091416-035426 |
| [2] |
D.C. Wallace, Nat. Genet. 50 (2018) 1642-1649. DOI:10.1038/s41588-018-0264-z |
| [3] |
H.M. McBride, M. Neuspiel, S. Wasiak, Curr. Biol. 16 (2006) R551-R560. DOI:10.1016/j.cub.2006.06.054 |
| [4] |
N. Hance, M.I. Ekstrand, A. Trifunovic, Hum. Mol. Genet. 14 (2005) 1775-1783. DOI:10.1093/hmg/ddi184 |
| [5] |
N.G. Larsson, J. Wang, H. Wilhelmsson, et al., Nat. Genet. 18 (1998) 231-236. DOI:10.1038/ng0398-231 |
| [6] |
M. Mittal, M.R. Siddiqui, K. Tran, S.P. Reddy, A.B. Malik, Antioxid. Redox Signal. 20 (2014) 1126-1167. DOI:10.1089/ars.2012.5149 |
| [7] |
J. Zielonka, J. Joseph, A. Sikora, et al., Chem. Rev. 117 (2017) 10043-10120. DOI:10.1021/acs.chemrev.7b00042 |
| [8] |
N. Wiedemann, N. Pfanner, Annu. Rev. Biochem. 86 (2017) 685-714. DOI:10.1146/annurev-biochem-060815-014352 |
| [9] |
S. Mallick, S.J. Song, Y. Bae, J.S. Choi, Int. J. Biol. Macromol. 132 (2019) 451-460. DOI:10.1016/j.ijbiomac.2019.03.215 |
| [10] |
L.I. Lin, Y.F. Ko, Y.C. Ko, J.K. Lin, Oncology 55 (1998) 349-353. DOI:10.1159/000011876 |
| [11] |
F. Zhang, N.K. Altorki, J.R. Mestre, K. Subbaramaiah, A.J. Dannenberg, Carcinogenesis 20 (1999) 445-451. DOI:10.1093/carcin/20.3.445 |
| [12] |
X. Feng, W. Xu, X. Xu, et al., Sci. China Chem. 64 (2020) 293-301. |
| [13] |
C. Zheng, M. Li, J. Ding, Bio Integr. 2 (2021) 57-60. DOI:10.15212/bioi-2021-0016 |
| [14] |
P. Zheng, B. Ding, Z. Jiang, et al., Nano Lett. 21 (2021) 2088-2093. DOI:10.1021/acs.nanolett.0c04778 |
| [15] |
P. Zheng, B. Ding, R. Shi, et al., Adv. Mater. 33 (2021) e2007426. DOI:10.1002/adma.202007426 |
| [16] |
R. Vaishya, V. Khurana, S. Patel, A.K. Mitra, Expert Opin. Drug Deliv. 12 (2015) 415-440. DOI:10.1517/17425247.2015.961420 |
| [17] |
S. Sikder, V. Gote, M. Alshamrani, J. Sicotte, D. Pal, Expert Opin, Drug Deliv. 16 (2019) 1113-1131. |
| [18] |
P. Agarwal, I.D. Rupenthal, Drug Discov. Today 18 (2013) 337-349. DOI:10.1016/j.drudis.2013.01.013 |
| [19] |
C. Chen, Z. Yang, X. Tang, Med. Res. Rev. 38 (2018) 829-869. DOI:10.1002/med.21479 |
| [20] |
M.R. Dyer, P.L. Herrling, Mol. Ther. 1 (2000) 213-224. DOI:10.1006/mthe.2000.0044 |
| [21] |
T.T.L. Yu, P. Gupta, V. Ronfard, A.A. Vertes, Y. Bayon, Front. Bioeng. Biotechnol. 6 (2018) 130. DOI:10.3389/fbioe.2018.00130 |
| [22] |
C.C. Ma, Z.L. Wang, T. Xu, Z.Y. He, Y.Q. Wei, Biotechnol. Adv. 40 (2020) 107502. DOI:10.1016/j.biotechadv.2019.107502 |
| [23] |
K.H. Vousden, Cell 103 (2000) 691-694. DOI:10.1016/S0092-8674(00)00171-9 |
| [24] |
S. Reaz, M. Mossalam, A. Okal, C.S. Lim, Mol. Pharm. 10 (2013) 1350-1359. DOI:10.1021/mp300598k |
| [25] |
G. Palacios, U.M. Moll, Oncogene 25 (2006) 6133-6139. DOI:10.1038/sj.onc.1209641 |
| [26] |
F. Talos, O. Petrenko, P. Mena, U.M. Moll, Cancer Res. 65 (2005) 9971-9981. DOI:10.1158/0008-5472.CAN-05-1084 |
| [27] |
G. Palacios, H.C. Crawford, A. Vaseva, U.M. Moll, Cell Cycle 7 (2008) 2584-2590. DOI:10.4161/cc.7.16.6070 |
| [28] |
C. Zhang, Z. Liu, Y. Zheng, et al., Small 14 (2018) 1703306. DOI:10.1002/smll.201703306 |
| [29] |
C. Han, C. Zhang, T. Ma, et al., Acta Biomater. 77 (2018) 268-281. DOI:10.1016/j.actbio.2018.07.018 |
| [30] |
W. Li, P. Zhan, E. De Clercq, H. Lou, X. Liu, Prog. Polym. Sci. 38 (2013) 421-444. DOI:10.1016/j.progpolymsci.2012.07.006 |
| [31] |
S.R. Jean, M. Ahmed, E.K. Lei, S.P. Wisnovsky, S.O. Kelley, Acc. Chem. Res. 49 (2016) 1893-1902. DOI:10.1021/acs.accounts.6b00277 |
| [32] |
A. Heller, G. Brockhoff, A. Goepferich, Eur. J. Pharm. Biopharm. 82 (2012) 1-18. DOI:10.1016/j.ejpb.2012.05.014 |
| [33] |
S.S. Liew, X. Qin, J. Zhou, et al., Angew. Chem. Int. Ed. 60 (2020) 2232-2256. |
| [34] |
J. Zielonka, J. Joseph, A. Sikora, et al., Chem. Rev. 117 (2017) 10043-10120. DOI:10.1021/acs.chemrev.7b00042 |
| [35] |
P. Zheng, Y. Liu, J. Chen, et al., Chin. Chem. Lett. 31 (2020) 1178-1182. DOI:10.1016/j.cclet.2019.12.001 |
| [36] |
Z. Luo, Y. Dai, H. Gao, Acta Pharm. Sin. B 9 (2019) 1099-1112. DOI:10.1016/j.apsb.2019.06.004 |
| [37] |
W. Zhang, F. Wang, C. Hu, et al., Acta Pharm. Sin. B 10 (2020) 2037-2053. DOI:10.1016/j.apsb.2020.07.013 |
| [38] |
B. Calitree, D.J. Donnelly, J.J. Holt, et al., Organometallics 26 (2007) 6248-6257. DOI:10.1021/om700846m |
| [39] |
L. He, K.N. Wang, Y. Zheng, et al., Dalton Trans. 47 (2018) 6942-6953. DOI:10.1039/C8DT00783G |
| [40] |
G.Y. Pan, H.R. Jia, Y.X. Zhu, et al., ACS Appl. Nano Mater. 1 (2018) 2885-2897. DOI:10.1021/acsanm.8b00527 |
| [41] |
G.Y. Pan, H.R. Jia, Y.X. Zhu, et al., ACS Biomater. Sci. Eng. 3 (2017) 3596-3606. DOI:10.1021/acsbiomaterials.7b00480 |
| [42] |
C. Gui, E. Zhao, R.T.K. Kwok, et al., Chem. Sci. 8 (2017) 1822-1830. DOI:10.1039/C6SC04947H |
| [43] |
M. Jiang, R.T.K. Kwok, X. Li, et al., J. Mater. Chem. B 6 (2018) 2557-2565. DOI:10.1039/C7TB02609A |
| [44] |
E. Zhao, H. Deng, S. Chen, et al., Chem. Commun. 50 (2014) 14451-14454. DOI:10.1039/C4CC07128J |
| [45] |
E.J. Ngen, P. Rajaputra, Y. You, Bioorg. Med. Chem. 17 (2009) 6631-6640. DOI:10.1016/j.bmc.2009.07.074 |
| [46] |
Y. Cheng, Y. Ji, J. Control. Release 318 (2020) 38-49. DOI:10.1016/j.jconrel.2019.12.011 |
| [47] |
J.H. Kang, Y.T. Ko, Pharmaceutics 11 (2019) 423. DOI:10.3390/pharmaceutics11080423 |
| [48] |
J.B. Rothbard, T.C. Jessop, R.S. Lewis, B.A. Murray, P.A. Wender, J. Am. Chem. Soc. 126 (2004) 9506-9507. DOI:10.1021/ja0482536 |
| [49] |
B. Kovaćevič, Z.B. Maksic, Org. Lett. 3 (2001) 1523-1526. DOI:10.1021/ol0158415 |
| [50] |
L. Wang, W. Du, Z. Hu, et al., Angew. Chem. Int. Ed. 58 (2019) 14026-14043. DOI:10.1002/anie.201901061 |
| [51] |
Y. Koide, Y. Urano, S. Kenmoku, H. Kojima, T. Nagano, J. Am. Chem. Soc. 129 (2007) 10324-10325. DOI:10.1021/ja073220m |
| [52] |
D. Zhang, L. Wen, R. Huang, et al., Biomaterials 153 (2018) 14-26. DOI:10.1016/j.biomaterials.2017.10.034 |
| [53] |
F. Lin, Y.W. Bao, F.G. Wu, Molecules 23 (2018) 3016. DOI:10.3390/molecules23113016 |
| [54] |
Y. Chen, W. Ai, X. Guo, et al., Small 15 (2019) 1902352. DOI:10.1002/smll.201902352 |
| [55] |
J. Xu, J. Pan, X. Jiang, et al., Biosens. Bioelectron. 77 (2016) 725-732. DOI:10.1016/j.bios.2015.10.049 |
| [56] |
J. Yan, J. Chen, N. Zhang, et al., J. Mater. Chem. B 8 (2020) 492-503. DOI:10.1039/C9TB02266J |
| [57] |
H. Cheng, R.R. Zheng, G.L. Fan, et al., Biomaterials 188 (2019) 1-11. DOI:10.1016/j.biomaterials.2018.10.005 |
| [58] |
S. Leo, G. Szabadkai, R. Rizzuto, Ann. N. Y. Acad. Sci. 1147 (2008) 264-274. DOI:10.1196/annals.1427.019 |
| [59] |
X. Zhang, F. Ai, T. Sun, F. Wang, G. Zhu, Inorg. Chem. 55 (2016) 3872-3880. DOI:10.1021/acs.inorgchem.6b00020 |
| [60] |
H.H. Szeto, A.V. Birk, Clin. Pharmacol. Ther. 96 (2014) 672-683. DOI:10.1038/clpt.2014.174 |
| [61] |
B.R. Lee, K.T. Oh, Y.T. Oh, et al., Chem. Commun. 47 (2011) 3852-3854. DOI:10.1039/c0cc03590d |
| [62] |
L. Jiang, L. Li, X. He, et al., Biomaterials 52 (2015) 126-139. DOI:10.1016/j.biomaterials.2015.02.004 |
| [63] |
P. Lu, B.J. Bruno, M. Rabenau, C.S. Lim, J. Control. Release 240 (2016) 38-51. DOI:10.1016/j.jconrel.2015.10.023 |
| [64] |
K.J. Matissek, A. Okal, M. Mossalam, C.S. Lim, Pharm. Res. 31 (2014) 2503-2515. DOI:10.1007/s11095-014-1346-y |
| [65] |
M. Mossalam, K.J. Matissek, A. Okal, J.E. Constance, C.S. Lim, Mol. Pharm. 9 (2012) 1449-1458. DOI:10.1021/mp3000259 |
| [66] |
E. Perales-Clemente, P. Fernandez-Silva, R. Acin-Perez, A. Perez-Martos, J.A. Enriquez, Nucleic Acids Res. 39 (2011) 225-234. DOI:10.1093/nar/gkq769 |
| [67] |
M.M. Mkandawire, M. Lakatos, A. Springer, et al., Nanoscale 7 (2015) 10634-10640. DOI:10.1039/C5NR01483B |
| [68] |
M. Li, Z. Zhong, J. Zhu, et al., J. Biol. Chem. 285 (2010) 14871-14881. DOI:10.1074/jbc.M109.069591 |
| [69] |
J.E. Constance, S.D. Despres, A. Nishida, C.S. Lim, Pharm. Res. 29 (2012) 2317-2328. DOI:10.1007/s11095-012-0758-9 |
| [70] |
H. Raza, FEBS J. 278 (2011) 4243-4251. DOI:10.1111/j.1742-4658.2011.08358.x |
| [71] |
K. Bruns, N. Studtrucker, A. Sharma, et al., J. Biol. Chem. 282 (2007) 353-363. DOI:10.1074/jbc.M606494200 |
| [72] |
S. Iwata, M. Saynovits, T.A. Link, H. Michel, Structure 4 (1996) 567-579. DOI:10.1016/S0969-2126(96)00062-7 |
| [73] |
I. Meton, M. Egea, F. Fernandez, M.C. Eraso, I.V. Baanante, FEBS Lett. 566 (2004) 251-254. DOI:10.1016/j.febslet.2004.04.051 |
| [74] |
I.N. Shokolenko, M.F. Alexeyev, S.P. LeDoux, G.L. Wilson, DNA Repair 4 (2005) 511-518. DOI:10.1016/j.dnarep.2004.11.009 |
| [75] |
K. Takaya, Y. Higuchi, K. Kitamoto, M. Arioka, FEMS Microbiol. Lett. 301 (2009) 201-209. DOI:10.1111/j.1574-6968.2009.01818.x |
| [76] |
N.D. Marchenko, A. Zaika, U.M. Moll, J. Biol. Chem. 275 (2000) 16202-16212. DOI:10.1074/jbc.275.21.16202 |
| [77] |
Y. Shen, L. Liang, J. Zhang, et al., Sens. Actuators B 285 (2019) 84-91. DOI:10.1016/j.snb.2019.01.036 |
| [78] |
M.S. Chan, P.K. Lo, Small 10 (2014) 1255-1260. DOI:10.1002/smll.201302993 |
| [79] |
G.S. Yu, J. Han, K.S. Ko, J.S. Choi, Macromol. Res. 22 (2013) 42-46. |
| [80] |
A. Klimpel, I. Neundorf, J. Control. Release 291 (2018) 147-156. DOI:10.1016/j.jconrel.2018.10.029 |
| [81] |
M. Sibrian-Vazquez, I.V. Nesterova, T.J. Jensen, M.G. Vicente, Bioconjugate Chem. 19 (2008) 705-713. DOI:10.1021/bc700393u |
| [82] |
J. Li, K. Pu, Acc. Chem. Res. 53 (2020) 752-762. DOI:10.1021/acs.accounts.9b00569 |
| [83] |
A.A. Zubareva, T.S. Shcherbinina, V.P. Varlamov, E.V. Svirshchevskaya, Nanoscale 7 (2015) 7942-7952. DOI:10.1039/C5NR00327J |
| [84] |
X. Song, Y. Chen, G. Zhao, et al., Carbohydr. Polym. 231 (2020) 115689. DOI:10.1016/j.carbpol.2019.115689 |
| [85] |
A.D. Ellington, J.W. Szostak, Nature 339 (1990) 519-522. |
| [86] |
C. Tuerk, L. Gold, Science 249 (1990) 505-510. DOI:10.1126/science.2200121 |
| [87] |
D.E. Huizenga, J.W. Szostak, Biochemistry 34 (1995) 656-665. DOI:10.1021/bi00002a033 |
| [88] |
T. Hermann, D.J. Patel, Biochemistry 287 (2000) 820-825. |
| [89] |
P. Mallikaratchy, R.V. Stahelin, Z. Cao, W. Cho, W. Tan, Chem. Commun. 30 (2006) 3229-3231. |
| [90] |
D.A. Daniels, H. Chen, B.J. Hicke, K.M. Swiderek, L. Gold, Proc. Natl. Acad. Sci. U. S. A. 100 (2003) 15416-15421. DOI:10.1073/pnas.2136683100 |
| [91] |
P. Parekh, Z.W. Tang, P.C. Turner, R.W. Moyer, W.H. Tan, Anal. Chem. 82 (2010) 8642-8649. DOI:10.1021/ac101801j |
| [92] |
L.C. Bock, L.C. Griffin, J.A. Latham, E.H. Vermaas, J.J. Toole, Nature 355 (1992) 564-566. DOI:10.1038/355564a0 |
| [93] |
J. Hu, J. Wu, C. Li, et al., ChemBioChem 12 (2011) 424-430. DOI:10.1002/cbic.201000470 |
| [94] |
C.B. Chen, G.A. Chernis, V.Q. Hoang, R. Landgraf, Proc. Natl. Acad. Sci. U. S. A. 100 (2003) 9226-9231. DOI:10.1073/pnas.1332660100 |
| [95] |
A.D. Keefe, S. Pai, A. Ellington, Nat. Rev. Drug Discov. 9 (2010) 537-550. DOI:10.1038/nrd3141 |
| [96] |
K.T. Guo, R. SchAfer, A. Paul, et al., Stem Cells 24 (2006) 2220-2231. DOI:10.1634/stemcells.2006-0015 |
| [97] |
D. Shangguan, Y. Li, Z.W. Tang, et al., Proc. Natl. Acad. Sci. U. S. A. 103 (2006) 11838-11843. DOI:10.1073/pnas.0602615103 |
| [98] |
Z.W. Tang, D. Shangguan, K.M. Wang, et al., Anal. Chem. 79 (2007) 4900-4907. DOI:10.1021/ac070189y |
| [99] |
D.H. Shangguan, L. Meng, Z.H.C. Cao, et al., Anal. Chem. 80 (2008) 721-728. DOI:10.1021/ac701962v |
| [100] |
K. Sefah, Z.W. Tang, D.H. Shangguan, et al., Leukemia 23 (2009) 235-244. DOI:10.1038/leu.2008.335 |
| [101] |
H.W. Chen, C.D. Medley, K. Sefah, et al., ChemMedChem 3 (2008) 991-1001. DOI:10.1002/cmdc.200800030 |
| [102] |
A.T. Bayrac, K. Sefah, P. Parekh, et al., ACS Chem. Neurosci. 2 (2011) 175-181. DOI:10.1021/cn100114k |
| [103] |
D. van Simaeys, D. Lopez-Colon, K. Sefah, et al., PLoS One 5 (2010) e13770. DOI:10.1371/journal.pone.0013770 |
| [104] |
Z. Zhao, L. Xu, X. Shi, et al., Analyst 134 (2009) 1808-1814. DOI:10.1039/b904476k |
| [105] |
S. Chakraborty, Z.Y. Dlie, S. Chakraborty, et al., Mol. Ther. Nucleic Acids 20 (2020) 34-49. DOI:10.1016/j.omtn.2020.01.034 |
| [106] |
H. Sun, X. Zhu, P.Y. Lu, et al., Mol. Ther. Nucleic Acids 3 (2014) e182. DOI:10.1038/mtna.2014.32 |
| [107] |
C. Xu, X.Y. He, Y. Peng, et al., Anal. Chem. 92 (2019) 2088-2096. |
| [108] |
G. Zhu, X. Chen, Adv. Drug Deliv. Rev. 134 (2018) 65-78. DOI:10.1016/j.addr.2018.08.005 |
| [109] |
C. Fiore, M. Salvi, M. Palermo, et al., Biochim. Biophys. Acta 1658 (2004) 195-201. DOI:10.1016/j.bbabio.2004.05.012 |
| [110] |
X. Wu, L. Zhang, E. Gurley, et al., Hepatology 47 (2008) 1905-1915. DOI:10.1002/hep.22239 |
| [111] |
X. Qin, C. Yu, J. Wei, et al., Adv. Mater. 31 (2019) e1902791. DOI:10.1002/adma.201902791 |
| [112] |
M. Ferrari, Nat. Rev. Cancer 5 (2005) 161-171. DOI:10.1038/nrc1566 |
| [113] |
S.E. McNeil, Nat. Rev. Mater. 1 (2016) 16073. DOI:10.1038/natrevmats.2016.73 |
| [114] |
O.C. Farokhzad, R. Langer, Adv. Drug Deliv. Rev. 58 (2006) 1456-1459. DOI:10.1016/j.addr.2006.09.011 |
| [115] |
K. Riehemann, S.W. Schneider, T.A. Luger, et al., Angew. Chem. Int. Ed. 48 (2009) 872-897. DOI:10.1002/anie.200802585 |
| [116] |
E. Fattal, N. Tsapis, Clin. Transl. Imaging 2 (2014) 77-87. DOI:10.1007/s40336-014-0053-3 |
| [117] |
J. Panyam, V. Labhasetwar, Adv. Drug Deliv. Rev. 55 (2003) 329-347. DOI:10.1016/S0169-409X(02)00228-4 |
| [118] |
I. Brigger, C. Dubernet, P. Couvreur, Adv. Drug Deliv. Rev 64 (2012) 24-36. DOI:10.1016/j.addr.2012.09.006 |
| [119] |
Y. Xue, H. Bai, B. Peng, et al., Chem. Soc. Rev. 50 (2021) 4872-4931. DOI:10.1039/D0CS01061H |
| [120] |
N. Bertrand, J. Wu, X. Xu, N. Kamaly, O.C. Farokhzad, Adv. Drug Deliv. Rev. 66 (2014) 2-25. DOI:10.1016/j.addr.2013.11.009 |
| [121] |
F.X. Gu, R. Karnik, A.Z. Wang, et al., Nano Today 2 (2007) 14-21. |
| [122] |
E. Ruoslahti, S.N. Bhatia, M.J. Sailor, J. Cell Biol. 188 (2010) 759-768. DOI:10.1083/jcb.200910104 |
| [123] |
A. Wicki, D. Witzigmann, V. Balasubramanian, J. Huwyler, J. Control. Release 200 (2015) 138-157. DOI:10.1016/j.jconrel.2014.12.030 |
| [124] |
M. Bortolato, K. Chen, J.C. Shih, Adv. Drug Deliv. Rev. 60 (2008) 1527-1533. DOI:10.1016/j.addr.2008.06.002 |
| [125] |
N. Apostolova, V.M. Victor, Antioxid. Redox Signal. 22 (2015) 686-729. DOI:10.1089/ars.2014.5952 |
| [126] |
T. Govender, S. Stolnik, C.D. Xiong, et al., J. Control. Release 75 (2001) 249-258. DOI:10.1016/S0168-3659(01)00353-4 |
| [127] |
V.J. Adlam, J.C. Harrison, C.M. Porteous, et al., FASEB J. 19 (2005) 1088-1095. DOI:10.1096/fj.05-3718com |
| [128] |
F. Bruni, R.N. Lightowlers, Mol. Ther. 19 (2011) 1404-1405. DOI:10.1038/mt.2011.139 |
| [129] |
Z. Chen, X. Kang, Y. Wu, et al., Chem. Commun. 55 (2019) 4781-4784. DOI:10.1039/C9CC00531E |
| [130] |
M. Hibino, Y. Yamada, N. Fujishita, et al., J. Pharm. Sci. 108 (2019) 2668-2676. DOI:10.1016/j.xphs.2019.04.001 |
| [131] |
J. Wang, R. Zhu, X. Sun, et al., Int. J. Nanomed. 9 (2014) 3987-3998. |
| [132] |
C. Yue, Y. Yang, J. Song, et al., Nanoscale 9 (2017) 11103-11118. DOI:10.1039/C7NR02193C |
| [133] |
N. Li, C.X. Zhang, X.X. Wang, et al., Biomaterials 34 (2013) 3366-3380. DOI:10.1016/j.biomaterials.2013.01.055 |
| [134] |
Y.L. Lo, C.S. Wang, Y.C. Chen, et al., Pharmaceutics 12 (2020) 756. DOI:10.3390/pharmaceutics12080756 |
| [135] |
X. Gu, C. Shen, H. Li, E.M. Goldys, W. Deng, J. Nanobiotechnol. 18 (2020) 87. DOI:10.1186/s12951-020-00644-z |
| [136] |
J. Tuo, Y. Xie, J. Song, et al., J. Mater. Chem. B 4 (2016) 6856-6864. DOI:10.1039/C6TB01730D |
| [137] |
Y. Yamada, H. Akita, H. Kamiya, et al., Biochim. Biophys. Acta 1778 (2008) 423-432. DOI:10.1016/j.bbamem.2007.11.002 |
| [138] |
Y. Yamada, H. Akita, K. Kogure, H. Kamiya, H. Harashima, Mitochondrion 7 (2007) 63-71. DOI:10.1016/j.mito.2006.12.003 |
| [139] |
L. Chen, Q. Song, Y. Chen, et al., ACS Nano 14 (2020) 6636-6648. DOI:10.1021/acsnano.9b09186 |
| [140] |
J.B. Rothbard, S. Garlington, Q. Lin, et al., Nat. Med. 6 (2000) 1253-1257. DOI:10.1038/81359 |
| [141] |
P.A. Wender, D.J. Mitchell, K. Pattabiraman, et al., Proc. Natl. Acad. Sci. U. S. A. 97 (2000) 13003-13008. DOI:10.1073/pnas.97.24.13003 |
| [142] |
S. Futaki, T. Suzuki, W. Ohashi, et al., J. Biol. Chem. 276 (2001) 5836-5840. DOI:10.1074/jbc.M007540200 |
| [143] |
V. Geromel, A. Cao, D. Briane, et al., Antisense Nucleic Acid Drug Dev. 11 (2001) 175-180. DOI:10.1089/108729001300338708 |
| [144] |
R. Kedmi, N. Ben-Arie, D. Peer, Biomaterials 31 (2010) 6867-6875. DOI:10.1016/j.biomaterials.2010.05.027 |
| [145] |
P. Charoensit, S. Kawakami, Y. Higuchi, F. Yamashita, M. Hashida, Cancer Gene Ther. 17 (2010) 512-522. DOI:10.1038/cgt.2010.12 |
| [146] |
J. Meng, F. Guo, H. Xu, Sci. Rep. 6 (2016) 22390. DOI:10.1038/srep22390 |
| [147] |
V.P. Torchilin, Nat. Rev. Drug Discov. 4 (2005) 145-160. DOI:10.1038/nrd1632 |
| [148] |
H. Li, J. Li, X. He, et al., Chin. Chem. Lett. 30 (2019) 1083-1088. DOI:10.1016/j.cclet.2019.01.003 |
| [149] |
D.Y. Cho, H. Cho, K. Kwon, et al., Adv. Funct. Mater. 25 (2015) 5479-5491. DOI:10.1002/adfm.201501422 |
| [150] |
Y. Liu, H. Li, J. Xie, et al., Biomater. Sci. 5 (2017) 1022-1031. DOI:10.1039/C6BM00878J |
| [151] |
F. Selmin, G. Magri, C.G. Gennari, et al., J. Pept. Sci. 23 (2017) 182-188. DOI:10.1002/psc.2952 |
| [152] |
Y. Tan, X. Yang, S. Dai, et al., Polym. Chem. 10 (2019) 512-525. DOI:10.1039/C8PY01504J |
| [153] |
W. Wang, J. Liu, W. Feng, et al., Biomater. Sci. 7 (2019) 1052-1063. DOI:10.1039/C8BM01414K |
| [154] |
J. Xu, B. Yan, X. Du, et al., Polym. Chem. 10 (2019) 983-990. DOI:10.1039/C8PY01518J |
| [155] |
Y. Xu, S. Wang, H.F. Chan, et al., Int. J. Pharm. 522 (2017) 21-33. DOI:10.1016/j.ijpharm.2017.01.064 |
| [156] |
S. Bai, X. Zhang, X. Ma, et al., Biomater. Sci. 6 (2018) 3126-3138. DOI:10.1039/C8BM01020J |
| [157] |
B.Y. Liu, W.X. Wu, Y.H. Liu, et al., Polym. Chem. 8 (2017) 5982-5987. DOI:10.1039/C7PY01138E |
| [158] |
T.M. Brenza, S. Ghaisas, J.E.V. Ramirez, et al., Nanomedicine 13 (2017) 809-820. DOI:10.1016/j.nano.2016.10.004 |
| [159] |
S. Chakrabortty, M. Sison, Y. Wu, et al., Biomater. Sci. 5 (2017) 966-971. DOI:10.1039/C6BM00951D |
| [160] |
D.B. Cheng, X.H. Zhang, Y.J. Gao, et al., J. Am. Chem. Soc. 141 (2019) 7235-7239. DOI:10.1021/jacs.8b07727 |
| [161] |
Y. Cheng, D.Z. Liu, C.X. Zhang, et al., Nanomedicine 16 (2019) 236-249. DOI:10.1016/j.nano.2018.12.014 |
| [162] |
C. Dong, Z. Liu, J. Liu, et al., Adv. Healthc. Mater. 5 (2016) 2214-2226. DOI:10.1002/adhm.201600212 |
| [163] |
H. He, A.W. Cattran, T. Nguyen, A.L. Nieminen, P. Xu, Biomaterials 35 (2014) 9546-9553. DOI:10.1016/j.biomaterials.2014.08.004 |
| [164] |
A. Ishikita, T. Matoba, G. Ikeda, et al., J. Am. Heart Assoc. 5 (2016) 1-17. |
| [165] |
Y.K. Kim, M. Zhang, J.J. Lu, et al., J. Drug Target. 24 (2016) 457-467. DOI:10.3109/1061186X.2015.1087527 |
| [166] |
E.B. Ku, K.T. Oh, Y.S. Youn, E.S. Lee, Macromol. Res. 24 (2016) 634-639. DOI:10.1007/s13233-016-4090-9 |
| [167] |
H.Q. Li, W.L. Ye, M.L. Huan, et al., Nanomedicine 14 (2019) 1011-1032. DOI:10.2217/nnm-2018-0227 |
| [168] |
Y. Liu, Z. Zhou, X. Lin, et al., Biomacromolecules 20 (2019) 3755-3766. DOI:10.1021/acs.biomac.9b00800 |
| [169] |
S. Marrache, S. Dhar, Proc. Natl. Acad. Sci. U. S. A. 109 (2012) 16288-16293. DOI:10.1073/pnas.1210096109 |
| [170] |
T. Qi, B. Chen, Z. Wang, et al., Biomaterials 213 (2019) 119219. DOI:10.1016/j.biomaterials.2019.05.030 |
| [171] |
C. Ren, D. Li, Q. Zhou, X. Hu, Biomaterials 232 (2020) 119752. DOI:10.1016/j.biomaterials.2019.119752 |
| [172] |
R. Zhang, L. Zhang, H. Ran, et al., Biomater. Sci. 8 (2020) 4581-4594. DOI:10.1039/D0BM00408A |
| [173] |
Z. Wang, X. Kuang, J. Shi, W. Guo, H. Liu, Biomater. Sci. 5 (2017) 1800-1809. DOI:10.1039/C7BM00224F |
| [174] |
J. Xu, J.G. Shamul, H. Wang, et al., Adv. Healthc. Mater. 9 (2020) 2000181. DOI:10.1002/adhm.202000181 |
| [175] |
H. Yao, L. Cao, W. Zhao, et al., J. Nanopart. Res. 19 (2017) 349. DOI:10.1007/s11051-017-4030-4 |
| [176] |
B.F. Zhang, L. Xing, P.F. Cui, et al., Biomaterials 61 (2015) 178-189. DOI:10.1016/j.biomaterials.2015.05.027 |
| [177] |
H. Zhou, C. Fu, X. Chen, et al., Biomater. Sci. 6 (2018) 1535-1545. DOI:10.1039/C8BM00142A |
| [178] |
S. Zhou, Y. Sun, X. Kuang, et al., Eur. J. Pharm. Sci. 126 (2019) 59-68. DOI:10.1016/j.ejps.2018.04.027 |
| [179] |
H. Hatcher, R. Planalp, J. Cho, F.M. Torti, S.V. Torti, Cell. Mol. Life Sci. 65 (2008) 1631-1652. DOI:10.1007/s00018-008-7452-4 |
| [180] |
L.Y. Shi, L. Zhang, H. Li, et al., Pharmacol. Rep. 70 (2018) 1040-1046. DOI:10.1016/j.pharep.2018.05.006 |
| [181] |
R.K. Pathak, N. Kolishetti, S. Dhar, Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 7 (2015) 315-329. DOI:10.1002/wnan.1305 |
| [182] |
J. Ahn, B. Lee, Y. Choi, et al., J. Mater. Chem. B 6 (2018) 5698-5707. DOI:10.1039/C8TB01358F |
| [183] |
N. Henry, J. Clouet, C.Le Visage, et al., J. Mater. Chem. B 5 (2017) 2908-2920. DOI:10.1039/C7TB00332C |
| [184] |
P. Yuan, X. Mao, X. Wu, et al., Angew. Chem. Int. Ed. 58 (2019) 7657-7661. DOI:10.1002/anie.201901699 |
| [185] |
C.E. Shi, C.Q. You, L. Pan, Nanotechnology 30 (2019) 325102. DOI:10.1088/1361-6528/ab1367 |
| [186] |
L. Harini, S. Srivastava, G.P. Gnanakumar, et al., Oncotarget 10 (2019) 1193-1208. DOI:10.18632/oncotarget.26623 |
| [187] |
K.I. Lee, C.C. Su, K.M. Fang, et al., Sci. Rep. 10 (2020) 9928. DOI:10.1038/s41598-020-66644-z |
| [188] |
S. Liang, C. Sun, P. Yang, et al., Biomaterials 240 (2020) 119850. DOI:10.1016/j.biomaterials.2020.119850 |
| [189] |
A. Pandey, K. Singh, S. Subramanian, et al., Chem. Eng. J. 394 (2020) 124419. DOI:10.1016/j.cej.2020.124419 |
| [190] |
L. Wang, Y. Liu, W. Li, et al., Nano Lett. 11 (2011) 772-780. DOI:10.1021/nl103992v |
| [191] |
M. Wang, K. Wang, G. Deng, et al., Int. J. Nanomed. 15 (2020) 2287-2302. DOI:10.2147/IJN.S240301 |
| [192] |
Y. Wang, F. Yang, H.X. Zhang, et al., Cell Death Dis. 4 (2013) e783. DOI:10.1038/cddis.2013.314 |
| [193] |
S. Han, R. Deng, Q. Gu, et al., Nature 587 (2020) 594-599. DOI:10.1038/s41586-020-2932-2 |
| [194] |
X.J. Zhu, R.F. Li, L. Xu, et al., Small 15 (2019) 1803428. DOI:10.1002/smll.201803428 |
| [195] |
M.S. Chan, L.S. Liu, H.M. Leung, P.K. Lo, ACS Appl. Mater. Interfaces 9 (2017) 11780-11789. DOI:10.1021/acsami.6b15954 |
| [196] |
M. Guo, H.J. Xiang, Y. Wang, et al., Chem. Commun. 53 (2017) 3253-3256. DOI:10.1039/C7CC00670E |
| [197] |
S. Marrache, S. Dhar, Chem. Sci. 6 (2015) 1832-1845. DOI:10.1039/C4SC01963F |
| [198] |
L. Zhang, J. Xia, Q. Zhao, L. Liu, Z. Zhang, Small 6 (2010) 537-544. DOI:10.1002/smll.200901680 |
| [199] |
Z. Li, J.C. Barnes, A. Bosoy, J.F. Stoddart, J.I. Zink, Chem. Soc. Rev. 41 (2012) 2590-2605. DOI:10.1039/c1cs15246g |
| [200] |
Z. Bai, J. Wei, C. Yu, et al., J. Mater. Chem. B 7 (2019) 1209-1225. DOI:10.1039/C8TB02946F |
| [201] |
J.Y. Yhee, S. Lee, K. Kim, Nanoscale 6 (2014) 13383-13390. DOI:10.1039/C4NR04334K |
| [202] |
J. Li, T. Zhao, T. Chen, et al., Nanoscale 7 (2015) 7502-7519. DOI:10.1039/C5NR00857C |
| [203] |
J. Kim, C. Jo, W.G. Lim, et al., Adv. Mater. 30 (2018) 1707557. DOI:10.1002/adma.201707557 |
| [204] |
I. Cohen-Erez, H. Rapaport, Colloids Surf. B (2018) 186-192. |
| [205] |
I. Cohen-Erez, N. Harduf, H. Rapaport, Polym. Adv. Technol. 30 (2019) 2506-2514. DOI:10.1002/pat.4707 |
| [206] |
S. Kim, H.Y. Nam, J. Lee, J. Seo, Biochemistry 59 (2020) 270-284. DOI:10.1021/acs.biochem.9b00857 |
| [207] |
E.K. Lei, S.O. Kelley, J. Am. Chem. Soc. 139 (2017) 9455-9458. DOI:10.1021/jacs.7b04415 |
| [208] |
I. Cohen-Erez, H. Rapaport, Biomacromolecules 16 (2015) 3827-3835. DOI:10.1021/acs.biomac.5b01140 |
| [209] |
X. Ma, X. Wang, M. Zhou, H. Fei, Adv. Healthc. Mater. 2 (2013) 1638-1643. DOI:10.1002/adhm.201300037 |
| [210] |
S.S. Malhi, R.S. Ramachandra Murthy, Drug Deliv. 9 (2012) 909-935. |
| [211] |
Y. Sun, A. Zhan, S. Zhou, et al., Chin. Chem. Lett. 30 (2019) 1435-1439. DOI:10.1016/j.cclet.2019.05.001 |
| [212] |
G.S. Price, R.L. Page, J.E. Riviere, J.M. Cline, D.E. Thrall, Cancer Chemother. Pharmacol. 38 (1995) 129-135. |
| [213] |
B. Silvestrini, G.M. Hahn, V. Cioli, C. De Martino, Br. J. Cancer 47 (1983) 221-231. DOI:10.1038/bjc.1983.30 |
| [214] |
G. Chen, I. Roy, C. Yang, P.N. Prasad, Chem. Rev. 116 (2016) 2826-2885. DOI:10.1021/acs.chemrev.5b00148 |
| [215] |
A.O. Elzoghby, W.M. Samy, N.A. Elgindy, J. Control. Release 161 (2012) 38-49190. DOI:10.1016/j.jconrel.2012.04.036 |
| [216] |
J. Li, T. Zhao, T. Chen, et al., Nanoscale 7 (2015) 7502-7519. DOI:10.1039/C5NR00857C |
| [217] |
D. Yang, B.C. Gates, ACS Catal. 9 (2019) 1779-1798. DOI:10.1021/acscatal.8b04515 |
| [218] |
Q. Wang, D. Astruc, Chem. Rev. 120 (2020) 1438-1511. DOI:10.1021/acs.chemrev.9b00223 |
| [219] |
S. Abednatanzi, P.G. Derakhshandeh, H. Depauw, et al., Chem. Soc. Rev. 48 (2019) 2535-2565. DOI:10.1039/C8CS00337H |
| [220] |
A. Terzopoulou, J.D. Nicholas, X.Z. Chen, et al., Chem. Rev. 120 (2020) 11175-11193. DOI:10.1021/acs.chemrev.0c00535 |
| [221] |
E. Doustkhah, R. Hassandoost, A. Khataee, R. Luque, M.H.N. Assadi, Chem. Soc. Rev. 50 (2021) 2927-2953. DOI:10.1039/C9CS00813F |
| [222] |
U. Kokcam-Demir, A. Goldman, L. Esrafili, et al., Chem. Soc. Rev. 49 (2020) 2751-2798. DOI:10.1039/C9CS00609E |
| [223] |
F. Saraci, V. Quezada-Novoa, P.R. Donnarumma, A.J. Howarth, Chem. Soc. Rev. 49 (2020) 7949-7977. DOI:10.1039/D0CS00292E |
| [224] |
P. Horcajada, T. Chalati, C. Serre, et al., Nat. Mater. 9 (2010) 172-178. DOI:10.1038/nmat2608 |
| [225] |
T.T. Chen, J.T. Yi, Y.Y. Zhao, X. Chu, J. Am. Chem. Soc. 140 (2018) 9912-9920. DOI:10.1021/jacs.8b04457 |
| [226] |
D. Liu, F. Jin, G. Shu, et al., Biomaterials 211 (2019) 57-67. DOI:10.1016/j.biomaterials.2019.04.034 |
| [227] |
M.C. Avendano-Monsalve, J.C. Ponce-Rojas, S. Funes, Biol. Chem. 401 (2020) 645-661. DOI:10.1515/hsz-2020-0110 |
| [228] |
Y. Huang, Z. Xiao, Z. Guan, et al., J. Control. Release 319 (2020) 119-134. DOI:10.1016/j.jconrel.2019.12.043 |
| [229] |
G.A. Kubi, Z. Qian, S. Amiar, et al., Angew. Chem. Int. Ed. 57 (2018) 17183-17188. DOI:10.1002/anie.201811940 |
| [230] |
H. Wang, F. Zhang, H. Wen, et al., J. Nanobiotechnol. 18 (2020) 8. DOI:10.1186/s12951-019-0562-3 |
| [231] |
X.W. Hua, Y.W. Bao, Z. Chen, F.G. Wu, Nanoscale 9 (2017) 10948-10960. DOI:10.1039/C7NR03658B |
| [232] |
R.X. Zhang, L.Y. Li, J. Li, et al., Adv. Funct. Mater. 27 (2017) 1700804. DOI:10.1002/adfm.201700804 |
| [233] |
R.A.J. Smith, C.M. Porteous, A.M. Gane, M.P. Murphy, Proc. Natl. Acad. Sci. U. S. A. 100 (2003) 5407-5412. DOI:10.1073/pnas.0931245100 |
| [234] |
S. Haddad, I.A. Lazaro, M. Fantham, et al., J. Am. Chem. Soc. 142 (2020) 6661-6674. DOI:10.1021/jacs.0c00188 |
| [235] |
K. Ni, G. Lan, S.S. Veroneau, et al., Nat. Commun. 9 (2018) 4321. DOI:10.1038/s41467-018-06655-7 |
| [236] |
A. Mallick, P. More, S. Ghosh, et al., ACS Appl. Mater. Interfaces 7 (2015) 7584-7598. DOI:10.1021/am5090226 |
| [237] |
J.H. Kang, Y.T. Ko, Biomater. Sci. 7 (2019) 2812-2825. DOI:10.1039/C9BM00403C |
| [238] |
L. Zhou, Y. Wu, X. Meng, et al., Small 14 (2018) 1801008. DOI:10.1002/smll.201801008 |
| [239] |
H. Zheng, L.J. Mortensen, S. Ravichandran, K. Bentley, L.A. DeLouise, J. Biomed. Nanotechnol. 13 (2017) 155-166. DOI:10.1166/jbn.2017.2337 |
| [240] |
S. Marrache, S. Tundup, D.A. Harn, S. Dhar, Methods Mol. Biol. 1265 (2015) 113-122. |
| [241] |
Q. Wang, J. Xu, R. Geng, et al., Biomaterials 231 (2020) 119671. DOI:10.1016/j.biomaterials.2019.119671 |
| [242] |
H.J. Liu, Y. Qin, Z.H. Zhao, et al., Theranostics 10 (2020) 9083-9099. DOI:10.7150/thno.46467 |
| [243] |
B. Banik, B.W. Askins, S. Dhar, Nanoscale 8 (2016) 19581-19591. DOI:10.1039/C6NR05882E |
| [244] |
H. Xiong, S. Du, J. Ni, J. Zhou, J. Yao, Biomaterials 94 (2016) 70-83. DOI:10.1016/j.biomaterials.2016.04.004 |
| [245] |
S.Y. Lee, H.J. Cho, Biomacromolecules 20 (2019) 835-845. DOI:10.1021/acs.biomac.8b01449 |
| [246] |
D. Boison, W.L. Lu, Q.M. Xu, et al., Colloids Surf. B 147 (2016) 387-396. DOI:10.1016/j.colsurfb.2016.08.024 |
| [247] |
Y. Lin, J. Liu, R. Bai, et al., ACS Nano 14 (2020) 11846-11859. DOI:10.1021/acsnano.0c04727 |
| [248] |
L. Liao, F. Jin, J. Nanosci. Nanotechnol. 16 (2016) 6690-6696. DOI:10.1166/jnn.2016.11375 |
| [249] |
K.Y. Kim, H. Jin, J. Park, et al., Nano Res. 11 (2017) 1082-1098. |
| [250] |
R. Wen, A.C. Umeano, L. Francis, et al., Vaccines 4 (2016) 18. DOI:10.3390/vaccines4020018 |
| [251] |
H. Chen, C. He, T. Chen, X. Xue, Biomater. Sci. 8 (2020) 3994-4002. DOI:10.1039/D0BM00500B |
| [252] |
L. Kong, L. Yang, C.Q. Xin, et al., Biosens. Bioelectron. 108 (2018) 14-19. DOI:10.1016/j.bios.2018.02.028 |
| [253] |
Y. Oh, J.Y. Je, M.S. Moorthy, H. Seo, W.H. Cho, Int. J. Pharm. 531 (2017) 1-13. DOI:10.1016/j.ijpharm.2017.07.014 |
| [254] |
J. Qu, D. Teng, G. Sui, et al., Biomater. Sci. 8 (2020) 3116-3129. DOI:10.1039/D0BM00003E |
| [255] |
X.H. Wang, H.S. Peng, L. Yang, et al., J. Mater. Chem. B 1 (2013) 5143-5152. DOI:10.1039/c3tb20884b |
| [256] |
E.I. Hassanen, A.F. Tohamy, M.Y. Issa, et al., Int. J. Nanomedicine 14 (2019) 8905-8922. DOI:10.2147/IJN.S229461 |
| [257] |
G. Yang, L. Xu, J. Xu, et al., Nano Lett. 18 (2018) 2475-2484. DOI:10.1021/acs.nanolett.8b00040 |
| [258] |
M.P. Murphy, R.A. Smith, Annu. Rev. Pharmacol. Toxicol. 47 (2007) 629-656. DOI:10.1146/annurev.pharmtox.47.120505.105110 |
| [259] |
S. Fulda, L. Galluzzi, G. Kroemer, Nat. Rev. Drug Discovery 9 (2010) 447-464. DOI:10.1038/nrd3137 |
| [260] |
C. Kirkby, E. Ghasroddashti, Med. Phys. 42 (2015) 1119-1128. DOI:10.1118/1.4906192 |
| [261] |
Y. Liu, J. Zhang, C. Zuo, et al., Nano Res. 9 (2016) 3257-3266. DOI:10.1007/s12274-016-1204-9 |
| [262] |
Y. Deng, F. Jia, X. Chen, Q. Jin, J. Ji, Small 16 (2020) 2001747. DOI:10.1002/smll.202001747 |
| [263] |
Y. Zheng, X. Ji, B. Yu, et al., Nat. Chem. 10 (2018) 787-794. DOI:10.1038/s41557-018-0055-2 |
| [264] |
Z. Shi, Q. Li, L. Mei, Chin. Chem. Lett. 31 (2020) 1345-1356. DOI:10.1016/j.cclet.2020.03.001 |
| [265] |
D. Wang, H. Huang, M. Zhou, et al., Chem. Commun. 55 (2019) 4051-4054. DOI:10.1039/C9CC00603F |
| [266] |
W. Yu, M. Shevtsov, X. Chen, H. Gao, Chin. Chem. Lett. 31 (2020) 1366-1374. DOI:10.1016/j.cclet.2020.02.036 |
| [267] |
Y. Zhou, H. Li, Y.W. Yang, Chin. Chem. Lett. 26 (2015) 825-828. DOI:10.1016/j.cclet.2015.01.038 |
| [268] |
L. Wang, L.L. Li, H.L. Ma, H. Wang, Chin. Chem. Lett. 24 (2013) 351-358. DOI:10.1016/j.cclet.2013.03.018 |
| [269] |
S. Li, Y. Gao, Y. Ding, A. Xu, H. Tan, Chin. Chem. Lett. 32 (2021) 313-318. DOI:10.1016/j.cclet.2020.04.049 |
| [270] |
Y. Li, Y. Song, W. Zhang, et al., J. Mater. Chem. B 8 (2020) 7382-7389. DOI:10.1039/D0TB01330G |
| [271] |
Y.M. He, L. Lei, J. Cao, et al., Sci. Adv. 7 (2021) 1-13. |
| [272] |
Y. Zhang, A. Khalique, X. Du, et al., Adv. Mater. 33 (2021) e2006570. DOI:10.1002/adma.202006570 |
| [273] |
X. Wang, S. Li, H. Liu, Photodiagnosis Photodyn. Ther. 34 (2021) 102218. DOI:10.1016/j.pdpdt.2021.102218 |
| [274] |
W. Park, S.J. Kim, P. Cheresh, et al., Biomater. Sci. 9 (2021) 5497-5507. DOI:10.1039/D1BM00419K |
| [275] |
H. Ren, Q. Yang, J. Yong, et al., Biomater. Sci. 9 (2021) 2709-2720. DOI:10.1039/D0BM02028A |
| [276] |
T. Zuo, J. Zhang, J. Yang, et al., Biomater. Sci. 9 (2021) 1872-1885. DOI:10.1039/D0BM01878C |
| [277] |
X. Di, D. Wang, J. Zhou, et al., Nano Lett. 21 (2021) 1651-1658. DOI:10.1021/acs.nanolett.0c04281 |
| [278] |
J. Xia, M. Qian, Q. Yao, et al., J. Control. Release 334 (2021) 263-274. DOI:10.1016/j.jconrel.2021.04.031 |
| [279] |
J. Xue, D. Duosiken, S. Zhong, et al., Acta Biomater. 131 (2021) 508-518. DOI:10.1016/j.actbio.2021.06.041 |
| [280] |
J. Yang, H. Zou, J. Ding, X. Chen, Acta Polym. Sin. 52 (2021) 960-977. |
 2022, Vol. 33
2022, Vol. 33 

