It is well known that the cell is the most basic unit of the morphological structure and life-sustaining activities of an organism. The birth, aging, disease and death of organisms are closely related to the gene expression of the corresponding cytokines. Therefore, understanding the composition, structure and function of cells, exploring the life activities of cells and studying the interaction between cells are of great significance for human cognition and control of the life activities of organisms [1]. It is clear that the analysis of biomacromolecules such as proteins, cytokines, exosomes, RNA and other substances in different single cells can give us some important clues about the cell, which is the key to understanding the role of cells. The single-cell analysis technology has matured in recent years [2, 3] and it has brought us many breakthrough achievements [4-10]. However, the analysis of single cells is proceeded based on the ability to effectively separate the target cells from heterogeneous cell samples. Single-cell separation is necessary and difficult since it is need to obtain pure single cells with very low concentration. A deeper understanding of the activity mechanism of partial cells remains a serious challenge [11]. Therefore, it is urgent to explore rapid, accurate, non-invasive and convenient methods for cell separation and subsequent analysis.
As a promising new emerging technology, the microfluidic chip has also shown its unique charm and played a crucial role in the culture, screening, enrichment and analysis of different types of cells [12-19]. In the past few years, these studies provided an important basis for understanding the growth mechanism, interaction and pathological mechanism of cells [16, 18-21]. Cell-related biological and physical properties are the key to distinguishing and screening different types of cells [22]. At the same time, microfluidic chip also provides a good toxicity evaluation platform for drug therapy and further promotes clinical application [15]. Usually, biological characteristics of cells, such as antigen-antibody specific binding, can be used to accurately identify and capture targets. On the other hand, specific microstructures can be designed in the microfluidic channel to screen and separate the target objects according to the physical properties of the cell such as size, charge performance, density and morphology. Such cells, separated by the physical properties of the microchannels, retain their original information because they are not affected by any chemical composition or physically damaged [23-25]. Therefore, physical separation is of great significance in the academic fields related to cell research.
Based on the important potential value of microfluidic technology in cell separation and subsequent analysis, this paper reviews the related research progress of microfluidic chips in recent years. The screening and analysis of cells using microfluidic chips can not only achieve a theoretical breakthrough, but also promote the early screening and diagnosis of diseases, the detection of drug therapy and the evaluation of treatment in clinical medicine. We believe that with the continuous improvement of microfluidic chips, cell-related information databases will be continuously filled, become conducive for targeted treatment of stubborn diseases and make great contributions to the future of human health.
2. Microfluidic methods for cell separationThe main methods of microfluidic chips used in cell screening, separation and enrichment can be roughly divided into the following two types: biochemical separation and physical separation [22]. The main principle of biochemical separation is the specific recognition of corresponding ligands by markers such as receptors or proteins on the cell surface membrane [26-29. In addition, physical separation depends on the different physical properties of different cells.
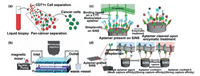
2.1. Biochemical separationThe biochemical purification and enrichment strategy on microfluidic chips is mainly conducted through combining cell surface antigens with specific ligands of strong affinity. Such ligands for capture are usually common antibodies or aptamers that are usually pre-modified on the surface of the microchannels or on the surface of the unit for post-processing collection. At present, it is a popular trend to capture circulating tumor cells (CTCs) by biochemical specific immunity. The reason is that CTCs related capture ligands are relatively common and easy to be obtained. The antibody for epithelial cell adhesion molecule (anti-EpCAM) is a common antibody used to capture cancer cells and has been widely used to detect cancer cells. It targets transmembrane glycoproteins on the surface of cancer cells [30]. In addition to the most commonly used affinity targets, some cancer cells have unique antigens that can also be captured as affinity targets, such as prostate-specific membrane antigen (PSMA) for prostate cancer [31] and epidermal growth factor receptor 2 (HER2) for breast cancer [32]. A number of anti-EpCAM-derived strategies have been investigated for capturing CTCs for related studies [33-36]. Studies have shown that CTCs can be captured by uniformly coating anti-EpCAM on the surface of the microfluidic channels [26]. However, there are many subtypes of cancer are missed in screening. New affinity ligands are key to solve this problem. Lyons et al. [26] found that human transferrin receptor (CD71) were demonstrated to be an affinity target for tumor cells in blood samples. As shown in Fig. 1a, an antibody named anti-CD71 was modified on the microfluidic channel to capture cancer cells with large amounts of CD71 on the surface. The results showed that six kinds of cancer cells could be captured by using CD71 as an affinity target and the capture purity > 80%, which indicated that CD71 had the potential to be used as an affinity marker to capture cancer cells. Different antigens used as affinity targets are specific to different antibodies leading to different capture efficiency and purity. Andree et al. [27] targeted EpCAM and HER2 as affinity antigens and applied several different antibodies on the microfluidic device shown in Fig. 1b to study the capture rate of cancer cells. The tested antibodies contain the EpCAM recognition antibodies HO-3 and VU1D9, and the HER2 recognition antibody HER81. The results showed that VU1D9 exhibited the best performance in capturing cancer cells compared with other antibodies and the best capturing rate was 0.6 mm/s. The results demonstrated that combining multiple antibodies on the capture surface may facilitate the capture of cells.
|
Download:
|
| Fig. 1. In situ biochemical separation on microfluidic chips. (a) Capturing cancer cells via human transferrin receptor. Copied with permission [26]. Copyright 2019, Elsevier B.V. (b) Anti-EpCAM body modified microfluidic chip for isolating CTCs. Copied with permission [27]. Copyright 2019, the Royal Society of Chemistry. (c) Aptamer present on SiNS for capturing target cells. Copied with permission [41]. Copyright 2014, American Chemical Society. (d) A novel "aptamer cocktail" for the precise enrichment of subspecies cells. Copied with permission [42]. Copyright 2016, Wiley-VCH GmbH. | |
Compared with antibodies, aptamers and peptides have more stable physical and chemical properties and are easy to be synthesized, so they can be used to replace antibodies in specific cell capture [37-39]. Aptamers functionalized with terminal groups, such as amino, carboxyl or sulfhydryl groups, can be easily self-assembled onto the substrate of the microfluidic channels. However, coating the antibodies or aptamers on the substrate of the microchannels is not satisfactory, because only the cells on the laminar surface in contact with the antibodies or aptamers can be recognized and trapped. In order to solve the problem of capture efficiency, the researchers changed the channel structure to achieve sufficient contact between the cells in the fluid and the capture ligands. Chen et al. [40] specifically developed a microfluidic platform consisted of microwell arrays which were encoded with cell-recognizable aptamer to isolate single tumor cells. Strong 3D local topographic interactions between the surface of target tumor cells and biomolecules were realized to obtain satisfied single-cell occupancy and unique bioselectivity. Moreover, nanostructures embedded in microchannels as aptamer carriers have been found to have an unexpected capture effect. Lin et al. [41] assembled the silicon nanowire substrate (SiNS) on the microchannels as the attachment points of aptamers. As shown in Fig. 1c, a silicon nanowire was embedded with a large number of aptamers, which greatly increased the contact area between the capture interface and fluid. More importantly, the dense nanofibers on the channel surface could cause Velcro effect between the captured cells and the SiNS surface to increase the binding force. Finally, the cells were stripped from the nanofibers by enzymes and obtained for downstream detection. It was undoubted that nanofibers had increased the efficiency of cell capture, but the accurate and effective capture of different subspecies of cells remained to be discussed. In order to improve the capture accuracy, Zhao et al. [42] proposed a reasonably designed "aptamer cocktail" with synergistic effect. As shown in Fig. 1d, the author selected different aptamers which showed different specificity for different CTC subtypes. Combining different aptamers on the SiNS could enhance the capture affinity of different cells through multiple receptor-ligand recognition. This study is of great significance for the precise enrichment of subspecies cells. But, it is difficult to find specific aptamers for capturing cells. It is currently available in vitro through systematically evolving ligands by exponential enrichment (SELEX) process [37-39]. However, the SELEX process is particularly time consuming and labor intensive. Lin et al. [43] recently assembled a system for automatically optimizing the screening of aptamers on microfluidic chip. The aptamer "H-45" with high specific affinity for ovarian cancer tissue samples was successfully obtained through a series of experiments. This means that the system has the potential to be a tool for screening specific ligands for cells that lack the recognition of ligands.
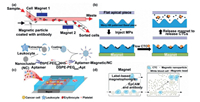
In addition to the above method of in situ capture, it is also a good choice to modify ligands on the surface of magnetic beads to capture target cells, because magnetic beads are easy to be modified and the separation of cells in microfluidic device has the advantages of high efficiency, simplicity and biocompatibility. Hejazian et al. [44] also summarized the physical laws involved in magnetic separation in microfluidic fluid, which provided the corresponding design reference for optimizing magnetic separation and purification. As shown in Fig. 2a, with the help of an external magnetic field, the magnetic beads can be precisely controlled to easily enrich the target cells [44]. Shi et al. [45] combined anti-EpCAM with magnetic particles (MPs), applied a magnetic field to the outside of the wave-shaped microfluidic chip as shown in Fig. 2b, and fixed the magnetic beads on the surface of the wave-shaped channel for capturing cells. After capturing, the magnetic field was removed to collect cells. It is necessary to design the anti-background interference of magnetic beads in order to improve the capture efficiency of target cells and prevent the non-specific adsorption of surrounding background cells. Zhang et al. [46] used white blood cell (WBC) membrane fragments to camouflage the surface of the beads, which inhibited the non-specific binding of WBCs to the beads. On this basis, the SYL3C aptamer was modified to capture EpCAM antigen on CTC membrane surface (Fig. 2c). The results showed that nearly 90% or more rare CTCs could be captured from whole blood within 20 min, with no nonspecific adsorption of WBCs. Although it is easy to capture, screen and enrich the cells with magnetic bead labeling, further processing is required for subsequent analysis and detection. To solve this problem, Zhao et al. [47] developed a label-free hydrodynamic cell separation (FCS) method for CTC isolation (Fig. 2d). The principle of the FCS method is that cells in ferrofluid flow are subjected to magnetic buoyance force, which varies according to the size of the cells. The results showed that FCS method had a high recovery rate and excellent biocompatibility, and was expected to be a powerful method for cell separation, and could be further combined with other methods to optimize the enrichment of cells. Hitherto, immunomagnetic enrichment of CTC cells has been widely used and yielded some exciting results [44-46, 48-50].
|
Download:
|
| Fig. 2. Magnetic beads used for capture cells. (a) Magnetic sorting cells in flowing fluid. Copied with permission [44]. Copyright 2015, the Royal Society of Chemistry. (b) Magnetic sorting cells in wave-shape microchannel. Copied with permission [45]. Copyright 2017, the Royal Society of Chemistry. (c) Leukocyte fragments modified beads mask nonspecific binding. Copied with permission [46]. Copyright 2019, American Chemical Society. (d) Label-free hydrodynamic cell separation. Copied with permission [47]. Copyright 2017, the Royal Society of Chemistry. | |
Physical separation is a method based on the combination of the physical characteristics of cells and the unique properties of microfluidic to achieve enrichment. In contrast to biochemical sorting that depends on the biological characteristics of the cell, primitive cells can be obtained by physical means without damage, eliminating the effects of ligand attachment, and providing a label-free enrichment platform for cell research. In recent years, scholars have invented many new sorting platforms based on different physical properties of cells, which have expanded new ideas for rapid screening.
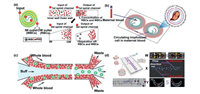
2.2.1. Pressure-driven hydrodynamic methodWhen a liquid flows in a spiral microchannel, Dean vortices are generated and emerge as two symmetrical counter-rotating vortices on the channel's cross section due to centrifugal acceleration. The researchers found that the inertial lift force brought the larger substances close to the inner wall (IW) of the channel, whereas the smaller ones were subjected to Dean force close to the outer wall (OW) [23, 24, 51, 52]. As shown in Fig. 3a, Jeon et al. [23] developed a double spiral microfluidic device for fully automatic leukocyte separation by using this rule. The purity of WBCs obtained by this device was as high as 99.99%, and up to 400, 000 WBCs without damage or surface deformation could be obtained from human peripheral blood of 50 µL. In addition, the device could be completely manually operated without damage on the separation efficiency and speed. Such convenient design, coupled with a high degree of deployability, provides a friendly platform for resource-limited environments. Huang et al. [24] developed a helical microfluidic chip to isolate non-invasive circulating fetal trophoblasts in order to detect prenatal genetic diseases in the fetus. As shown in Fig. 3b, circulating fetal trophoblasts carrying complete genetic information were collected at the branch near the inner wall of the channel under the action of inertial and Dean forces, providing a label-free, low-cost and non-invasive cell capture platform for prenatal assessment. There are many other examples of cell sorting using helical microchannels, but most of them are common and greatly different in target size. Most of the studies on the use of spiral microchannel separation and purification are limited to the separation of CTCs or other macromolecules from whole blood with large difference in the size [52-56]. However, the focusing effect of this method on cells of similar size is not obvious, and the separation of cells with small size difference still needs to be further optimized in the conditions of microchannels and adjusting the parameters. In order to solve this problem, it is necessary to further explore the influencing factors of spiral microchannel cell sorting. Exploring the focusing behavior of physical parameters other than size under the action of Dean vortices and inertial lift may be an effective way to solve this problem. Guzniczak et al. [57] conducted a theoretical study on the physical quantities involved. The authors simulated cells of different sizes and different deformations under different Reynolds coefficients. The results showed that the cell deformation had a great effect on the location of the cells under a sufficiently large Reynolds coefficient. This study made up for the deficiency of relevant theoretical data and influencing factors and made a great contribution to improve the purity of separation.
|
Download:
|
| Fig. 3. Hydrodynamic methods for cell separation. (a) Double spiral microfluidic device for fully automatic leukocyte separation. Copied with permission [23]. Copyright 2020, the Royal Society of Chemistry. (b) Helical microfluidic chip to isolate non-invasive circulating fetal trophoblasts. Copied with permission [24]. Copyright 2020, the Royal Society of Chemistry. (c) Shear-induced migration for the separation of WBCs. Copied with permission [25]. Copyright 2019, the Royal Society of Chemistry. (d) Multistep separation device. Copied with permission [58]. Copyright 2020, Elsevier B.V. | |
In addition, shear flow is also a common method which induces cells with different size to change the migration trajectory. When a fluid meets another fluid, a shear flow will be generated at the contact surface, and the larger material in the fluid will migrate to the shear flow generated at the contact surface and change the motion path. Using the principle of shear-induced migration, Zhou et al. [25] designed a microchannel as shown in Fig. 3c for the separation of WBCs from whole blood cells. The results showed that this method had the advantage of fast migration and provided a new simple method with high throughput for the isolation of rare cells. The cell-size-based hydrodynamic separation method used in above works was well with a small number of cell types, but as the number of cell types increased, unexpected things would happen. In order to solve this problem, different kinds of composite multifunctional microfluidic chips have been developed. Different kinds of cells were screened by different functions of microchannels to improve the separation accuracy. Gao et al. [58] designed a microfluidic device to isolate tumor cells from whole blood as shown in Fig. 3d. Briefly, the whole blood was injected through the inlet, firstly changing the distribution of different cells in the "W" region through the shear force generated by the chamber and the wall lift force. Then, the flow direction of most red blood cells (RBCs) was shifted with the help of the micropillars in the "X" region. Finally, large tumor cells were isolated from whole blood cells through an inertial focusing process of wavy microchannels. Mixed-separation devices also serve a number of other purposes [59-61]. Although the structure of the multi-stage separation system is complex, this kind of combined multi-step separation method can obtain relatively pure target cells and play an unexpected effect under the condition of strict requirements on the purity of the research object.
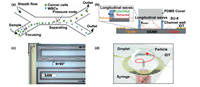
2.2.2. Acoustic separationWhen a standing ultrasonic wave is applied to the laminar flow perpendicular to the microchannel, the cells in the laminar flow will be subjected to the resonance force generated by the fluid and the ultrasonic wave. Due to the different physical characteristics of cell types and the ultrasonic force, cell sorting can be achieved with different sizes, densities and deformability. Wu et al. [62] integrated interdigital transducers (IDT) in microchannels to generate surface acoustic wave (SAW) for the isolation of rare tumor cells from WBCs (Fig. 4a). Moreover, Richard et al. [63] assembled a pair of IDTs on the microfluidic chip for generating standing surface acoustic wave (SSAW) in the microchannel. Fig. 4b shows the schematic diagram of the device generating SSAW in the microchannel. The author not only successfully enriched platelets from the whole blood, but also carried out a fully coupled three-dimensional (3D) numerical simulation of the data related to SAW wave field, and made a comprehensive analysis of the wave field generated in the channel. The experimental results were in good agreement with the simulation results, which made a theoretical contribution to the separation of cells by acoustic field method, and opened up a new possibility for the application of high-throughput cell separation. The physics of applying sound waves to non-linear microchannels can produce a curious phenomenon, which can be properly used to separate cells. Devendran et al. [64] obtained a time-average spatially varying landscape of sound pressure by applying a traveling wave in a serpentine microchannel. The traveling wave was caused by the diffraction effect of the sound field in the channel, so it was also called diffractive-acoustic SAW (DASAW). The schematic diagram of the DASAW device was shown in Fig. 4c. The simulation and optimization of the device showed that the focusing effect on continuous particles was excellent, and we speculated that it could be used for continuous separation of cells of different sizes. At present, almost all SAWs used for separation and purification are symmetric. To prove that asymmetric SAWs can be applied in particle separation, Zhang et al. [65] demonstrated a novel omnidirectional spiral surface acoustic wave (OSSAW) device which was used to separate RBCs and platelets from mouse blood with 93% and 84% purity, respectively (Fig. 4d). In order to obtain more accurate results for downstream analysis, strict cell sorting is required. Droplet capture of single cells can greatly reduce the risk of cross-contamination. At present, microfluidic droplet technology coupled with acoustic wave has been widely used in capture and analysis of single cells [66-74].
|
Download:
|
| Fig. 4. Acoustic cell separation on microfluidic chips. (a) SAW for the isolation of rare tumor cells. Copied with permission [62]. Copyright 2019, the Royal Society of Chemistry. (b) SSAW for the isolation of cells. Copied with permission [63]. Copyright 2019, the Royal Society of Chemistry. (C) Focusing effect of diffracted acoustic SAW devices on continuous particles. Copied with permission [64]. Copyright 2020, the Royal Society of Chemistry. (D) A novel OSSAW device for separation and purification. Copied with permission [65]. Copyright 2021, the Royal Society of Chemistry. | |
The method of cell separation based on electrophoresis is applying an electric field to the laminar flow in the microfluidic channel to induce the migration of cells with different charged properties in the fluid. The net charge on the surface of a cell is an important indicator that distinguishes a normal cell from a cancer cell, and some studies indicate that cancer cells have more negative charges than normal cells [75, 76]. Based on this knowledge, Jahangiri et al. [75] introduced a direct current electric field into the microfluidic chip (Fig. 5a), and successfully enriched a variety of cancer cells under the action of electric field force. Han et al. [77] proposed a technique for cell separation in a droplet, as shown in Fig. 5b, in which a pair of electrodes with different angles were assembled on a microfluidic chip to generate positive or negative dielectrophoresis forces on different cells. Different types of cells in the same droplet received different electrophoretic forces when they were passing through two pairs of electrodes, leading to the separation of the two types of cells which finally dispersed into two sub-droplets in the Y-shaped structure. Localizing the lateral position of a single cell in a microchannel is very important for cell focusing and sorting. Kung et al. [78] demonstrated a microfluidic device using tunnel dielectric electrophoresis (TDEP) to separate cells based on size differences (Fig. 5c). High resolution 3D operation of the spatial position of cells can be realized by the device, and the spatial position of cells of different sizes can be adjusted by regulating the division of electric field. This study provides a 3D focusing method with high spatial accuracy and provides a new idea for accurate cell sorting. Yang et al. [79] proposed an N-shaped electrode-based microfluidic impedance cytometer for the lateral location of a single cell in a fluid (Fig. 5d). The authors encoded the flow trajectories of individual particles by the differential current collected by N-type electrodes. According to the relationship between the position of flow particle, electrode, microchannel and current, a simple analytical formula was derived. This study provides a simple localization method for cell sorting. Due to its small size and easy integration, this device provides a quick and unlabeled test method for guaranteeing the accuracy of cell sorting. It has the potential to be used as a positioning system in highly integrated microfluidic chips. Huang et al. [80] presented a microfluidic chip with a non-uniform self-aligned sequential field, which enabled progressive drift of single-cell streamlines under dielectrophoresis. Isolated erythrocytes were evaluated with the help of a single-cell impedance cytometry. The results showed that the cells collected by the device were intact and this method could solve the problem of "all-electric" selective cell separation. This concept is expected to be used in cell detection systems for quantifying phenotypic heterogeneity of cellular systems. In general, this label-free cell manipulation method is easy to be integrated into the microfluidic chip, and the separation effect of particles with large charge difference is obvious. Furthermore, the accurate 3D position of the particles within the microchannel can improve the precision of the cell manipulation, so it has been widely used in particle and cell manipulation in microfluidics [77-83].
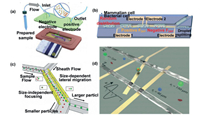
|
Download:
|
| Fig. 5. Electrophoresis methods for cell separation. (a) Electric field induced cell separation. Copied with permission [75]. Copyright 2019, Elsevier B.V. (b) Dielectrophoresis force induced different cell separation. Copied with permission [77]. Copyright 2020, the Royal Society of Chemistry. (c) TDEP for separation cells. Copied with permission [78]. Copyright 2021, the Royal Society of Chemistry. (d) N-shaped electrode-based microfluidic impedance cytometer for the lateral location of a single cell. Copied with permission [79]. Copyright 2019, the Royal Society of Chemistry. | |
According to the different shape and size of cells, it is also a common sorting method to prepare the corresponding size of traps or filter unit on the microfluidic chips [84-87]. Lin et al. [84] prepared a simple microfluidic chip for capturing cells. As shown in Fig. 6a, the channels on the microchip were distributed with traps similar to a sieve for intercepting cells of the corresponding size. In the meantime, gravity driven longitudinal filtration was introduced as another conducive way to increase flux and thus improve the capture efficiency. The microfluidic filter designed by Liu et al. [88] significantly improved the capture purity and throughput of cells. As shown in Fig. 6b, the author proposed a cell separation device with microporous array filtration. The device can quickly and sensitively isolate rare cells from a large number of samples. However, it is difficult to screen cells with similar size. In order to screen CTCs in whole blood, Ko et al. [89] changed the microporous filter membrane to be magnetic micropores, and modified magnetic markers on the surface of WBCs which were intercepted by magnetic micropores. Finally, a size-based capture device was used to separate RBCs and CTCs in the downstream. Yuan et al. [90] proposed a continuous sheathless separation method, which was verified by demonstrating separation of microalgal cells and bacteria. As shown in Fig. 6c, due to size-based differential viscoelastic focusing, the microalgae cells moved to the channel wall, while the bacteria moved to the channel centre. The authors pointed out that this method could also be applied to isolate animal cells, and this study provided a crucial method and idea for the separation of cells and bacteria.
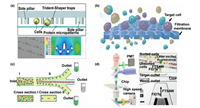
|
Download:
|
| Fig. 6. Other microfluidic method for cell separation. (a) Size-based traps for capturing cells. Copied with permission [84]. Copyright 2013, the Royal Society of Chemistry. (B) Gravity driven longitudinal filtration. Copied with permission [88]. Copyright 2019, the Royal Society of Chemistry. (C) Size-based differential viscoelastic focusing. Copied with permission [90]. Copyright 2019, the Royal Society of Chemistry. (D) Schematic illustration of FACS. FTSAW: focused traveling SAW. Copied with permission [91]. Copyright 2017, the Royal Society of Chemistry. | |
Fluorescence activated cell sorting (FACS) is an important method of cell screening, but FACS devices are very complex, bulky and expensive. In order to solve the shortcomings of common FACS devices, Ma et al. [91] integrated FACS into microfluidic chips. A schematic diagram of the device is shown in Fig. 6d. When the fluorescent signal generated by the laser excitation of the sorted cells was received by a photomultiplier tube (PMT), the focused interdigital transducer (FIDT) generated a focused acoustic beam to act on the target cell and changed its migration direction. In recent years, Raman activated cell sorting (RACS) has provided a high-throughput and non-invasive means for isolating cells. Single cell Raman spectra (SCRS) are intrinsic biochemical profiles and 'chemical images' of single cells which can be used to characterize phenotypic changes, physiological states and functions of cells [92]. By combining SCRS with 2D hydrodynamic focusing, Lyu et al. [93] developed a RACS system for continuously automatic sorting of single cells on a microfluidic chip. Hu et al. [94] proposed a new optical separation strategy. The optical constants of RBCs-conjugated CTCs (connecting CTCs with homologous RBC, CC-RBCs) were significantly different from those of other blood cells. When the modified CTCs flowed through the parallel infrared region, CC-RBCs could be separated from other cells precisely under the action of optical force. Magneto-driven cell screening has been described previously. Most studies have used magnetic beads to tag cells to indirectly drive screening of target cells [44-46]. The label-less FCS method demonstrated by Zhao et al. [47] provided a novel idea for magnetically driven separation. Through microfluidic assembly of mesoscopic superparamagnetic cores, a variety of stray magnetic field responses were constructed. The stray magnetic field fingerprint is recognized by the giant magnetoresistance sensor and can simultaneously parallel screen multiple targets, which provides an opportunity for magnetic multiplexing and enables the magnetic mixing, cleaning, concentration and separation of the analyte.
3. Subsequent cell analysis on microfluidic chipsAfter the capture of a specific target cell is completed, the cells need to be analyzed in order to further obtain the related biological information of the cell. Evaluation of cell counts and molecular characterization is key to understand cell-related physiological information, which helps us to improve our understanding of related cell biology. At present, the methods of cell analysis are relatively mature and diversified [95]. With the development of analytical tools, researchers are discovering that the information carried by single cells is important for current and future research [96-98].
3.1. Fluorescence analysisFluorescence detection is one of the most commonly used methods in the field of analysis at present. It has significant advantages such as high sensitivity, high resolution and simple operation. Moreover, the corresponding device has great compatibility with microfluidic chip and provides a convenient and efficient means for cell analysis [99-101]. Guo et al. [102] designed an ultra-sensitive platform for CTC detection based on the fluorescence characteristics of lanthanide nanoprobes. The surface of the nanoprobe is modified with anti-EpCAM antibody for the specific recognition of cancer cells (Fig. 7a). The device can realize the advantage of signal amplification by dissolution enhanced time-resolved photoluminescence (TRPL), and overcomes the problem of short life of common probes. The results showed that CTCs in whole blood could be detected directly, and the detection rate of CTCs in patients' blood was 93.9%. This study provides an effective early diagnostic strategy for rapid detection of cancer cells. Gallina et al. [103] developed a radiometric method for quantitative measurement of cells. As shown in Fig. 7b, radioactive labeled single cells and radioactive fluorescent probe were encapsulated in the same droplet. Fluorescent signal was produced due to the reactive oxygen species (ROS) produced by radioactive decomposition of water reacted with radioactive fluorescent probe. The experiment proved that the fluorescence signal was proportional to the radiation level. This experiment provides a new way for cell quantification. In order to study the activity of various proteases of cells, Ng et al. [104] prepared multi-color enzyme substrates based on fluorescence resonance energy transfer (FRET) and encased it in droplets along with the analytical cells. The schematic diagram of its mechanism is shown in the Fig. 7c. The protease secreted by cells acted on specific enzyme recognizer and emitted specific fluorescence. The expression activity of the protease could be identified by detecting the intensity of fluorescence. The authors used this device to measure the potential protease profiles expressed by different cancer cells, providing valuable insights into the study of cancer cells. A comprehensive understanding of cells requires the analysis of their basic functions which are controlled by miRNAs in the cells. Liu et al. [105] synthesized a plasma nanosensor functionalized with two nucleic acids in order to analyze miRNAs in cells (Fig. 7d). Silver nanoparticles (AgNPs) can enhance the fluorescence signal through plasma coupling before miRNA binding, but when miRNA binds to the target, the interference coupling reduces the fluorescence signal, thus accurately quantifying the concentration of the target miRNA in a single nucleus and cytoplasm. The authors used the microfluidic chip to accurately manipulate the cells. A single MCF-7 cell was wrapped in the same droplet with the functionalized nanosensor, and the cytoplasmic miR-155 and the nuclear miR-25 were simultaneously measured by fluorescence intensity. This study provides a new means to gain insight into the full range of biological processes at the subcellular level.
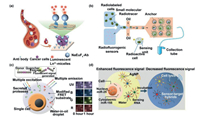
|
Download:
|
| Fig. 7. Fluorescent cell analysis on microfluidic chips. (a) Lanthanide nanoprobes for cell analysis. Copied with permission [102]. Copyright 2019, Wiley-VCH GmbH. (b) Radiometric method for quantitative measurement of cells. Copied with permission [103]. Copyright 2017, American Chemical Society. (c) FRET for detection of cells. Copied with permission [104]. Copyright 2016, Elsevier B.V. (d) Nucleic acid-functionalized plasma nanosensors for the analysis of miRNAs in cells. Copied with permission [105]. Copyright 2020, the Royal Society of Chemistry. | |
Raman spectroscopy has significant sensitivity and molecular fingerprint spectral characteristics, which has a great potential in single-cell analysis [92]. Single-cell Raman spectra based on microfluidic systems are gradually diversified. Various Raman flow cytometry techniques have been developed to address the shortcomings of traditional Raman analysis. The following mainly introduces several representative Raman derivative methods, such as surface-enhanced Raman scattering (SERS), multiplex coherent anti-Stokes Raman scattering (MCARs) and multiplex stimulated Raman scattering flow cytometry (SRS-FC).
SERS has been used to directly measure the cells captured by SERS nanoparticle probes. The advantage of this method is that Raman analysis only targets the cells labeled by SERS probes, and the unlabeled cells are not be affected even in the Raman region, so this analysis method does not need to enrich, separate and purify the cells. Wang et al. [106] designed a microfluidic Raman biochip, as shown in Fig. 8a, in which anti-63 magnetic nanoparticles were used to enrich exosomes. Exosomes were quickly captured by nanoparticles through triangular microcolumn array, and then EpCAM-functionalized Raman beads were introduced to couple with exosomes to form sandwich-like immune complexes, which was finally fixed in the Raman detection area with applied magnetic field for detection. The authors tested the device on patients with prostate cancer and compared the results with healthy samples. The results showed that the test procedure could be completed in less than 1 h and was able to successfully distinguish between the two samples, with a detection limit of 1.6 × 102 particles/mL. However, the detection method using the probe capture is usually influenced by the probe, resulting in biased information transmission. MCARs is a label-free nonlinear optical method for probing single cell and can obtain abundant molecular information by detecting the Raman energy within molecules. Camp et al. [107] developed the MCARs microfluidic flow cytometer shown in Fig. 8b, which excited multiple Raman vibrations simultaneously and had the ability to characterize thousands of samples per second. Conventional spontaneous Raman scattering flow cytometry has a slower rate. In order to achieve fast and high-throughput Raman analysis, Zhang et al. [108] proposed a technique called multiplex SRS-FC (Fig. 8c) which enabled rapid, label-free and accurate detection of single cell chemical composition. Spectral acquisition rates are four orders of magnitude higher than conventional Raman flow cytometry, with a throughput of up to 11, 000 particles per second. Under the cooperation with compositional principal component analysis (CPCA), the device detected a variety of chemical components of objects in 32 spectral channels. The author demonstrated that the device could detect the chemical components of 3T3-L1 cells with different differentiation states demonstrating the concept that the device could classify and detect different chemical components. This technique extends the approach of Raman analysis and provides new opportunities for high-throughput and multi-component analysis of cellular chemical components.
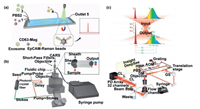
|
Download:
|
| Fig. 8. Raman cell analysis on microfluidic chips. (a) Schematic illustration of microfluidic Raman biochip. PBS: phosphate buffer saline. Copied with permission [106]. Copyright 2020, the Royal Society of Chemistry. (b) MCARs microfluidic flow cytometer. Copied with permission [107]. Copyright 2009, the Optical Society. (c) Schematic illustration of multiplex SRS-FC. HWP: half-wave plate; L: lens; AOM: acousto-optic modulator; GS: galvo scanner; DM: dichroic mirror; PBS: polarization beam splitter; CL: cylindrical lens; SP: short-pass filter. Copied with permission [108]. Copyright 2017, the Optical Society. | |
It is also important to analyze the morphology of cells, from which important information such as the growth environment and dynamic characteristics of cells can be determined. It is a convenient analytical method to directly obtain the visual morphological information of cells by microscopy. Wang et al. [109] demonstrated a high-throughput label-free microscopic hyperspectral imaging microchip for rapid evaluation of extracellular vesicles (EV) (Fig. 9a). EV membrane protein and corresponding antibodies have been successfully evaluated by using this device, which showed its value in the research of disease diagnosis and treatment. On the other hand, Siu et al. [110] developed a neural network assisted imaging flow cytometer (IFC) platform that allowed the analysis of the intrinsic morphological descriptors of the optical and mass density of a single cell in millions of cell populations (Fig. 9b). This deep learning assisted massive single-cell analysis strategy demonstrates the ability to delineate the biophysical characteristics of cancer subtypes without labels. This cutting-edge optical fluidic imaging analysis technique can be used not only to detect rare cell populations in heterogeneous samples, but also to evaluate the efficacy of targeted therapies. Multidirectional imaging flow cytometry (mIFC) is an upgraded version of the traditional IFC and can give 3D images of geometric features of a target. Kleiber et al. [111] cleverly took advantage of the fluid characteristics of microfluidic and integrated the flow rotation unit on the microchip, so that the target could be rotated so as to image it several times to obtain a 3D geometric image. Luo et al. [112] proposed a real-time quantitative phase microscope (RT-QPM) for 3D visualization of droplets in microfluidic. This technique can be used to measure the concentration distribution of analyte in droplet. It provides an effective means for droplet-based single cell analysis. The combination of this technology with the previously mentioned droplet-based technologies for single-cell capture [66-74, 113, 114] can achieve an integrated separation and analysis platform.

|
Download:
|
| Fig. 9. Cytomicroscopic imaging for cells on microfluidic chips. (a) Schematic representation of a high-throughput label-free microscopic hyperspectral imaging microchip. Copied with permission [109]. Copyright 2021, the Royal Society of Chemistry. (b) Neural network assisted IFC platform. Copied with permission [110]. Copyright 2020, the Royal Society of Chemistry. | |
Mass spectrometry (MS) has attracted much attention in the field of analysis due to its characteristics such as label-free, high detection limit, high efficiency and high sensitivity [14, 115-117]. In recent years, the technique has also shown unique advantages in single-cell analysis and obtained high-dimensional data from a single cell [118-122]. The combination of MS and microfluidic chips can provide a powerful multiple analysis tool with high throughput, high sensitivity and high selectivity for single-cell analysis [116, 117, 123-127]. According to the forms of ionization, MS can be divided into different types, including electrospray ionization mass spectrometry (ESI-MS), inductively coupled plasma mass spectrometry (ICP-MS), matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS). Different ionization modes lead to different coupling modes between the microfluidic chips and the mass spectrometers. The interface of the chip is important for the coupling of the chip with MS. Pedde et al. [128] summarized the methods of coupling ESI-MS and MALDI-MS with microfluidic chips. As shown in Fig. 10a, the different interface forms for ESI-MS and MALDI-MS are designed respectively. Different interface designs can achieve different purposes. For ESI-MS, the multiple emitter interfaces can simultaneously analyze multiple groups of target analytes, while the capillary emitter can accurately control the size of droplets entering into ESI from the microfluidic chip. For MALDI-MS, in addition to the droplet system-based dispensing device, the authors also introduce the capillary and integrated piezoelectric micro-dispensing which can realize automatic positioning. With the support of microfluidic technology, uniform, small and concentrated droplets of the target analyte sample can be automatically generated by any distribution method. Compared with standard pipetting technology, the sensitivity and reproductivity of MS can be improved.

|
Download:
|
| Fig. 10. MS analysis coupled with microfluidic chips. (a) Methods of coupling ESI-MS and MALDI-MS with microfluidic chips. Copied with permission [128]. Copyright 2017, Elsevier B.V. (b) Microextraction analysis of intracellular components. Copied with permission [129]. Copyright 2016, Springer Nature Limited. (c) Decoupled the droplet from the electric field via high volte. Copied with permission [132]. Copyright 2020, the Royal Society of Chemistry. (d) High-throughput MALDI-MS microfluidic platform for automatic analysis of single-cell phospholipids. Copied with permission [133]. Copyright 2015, American Chemical Society. | |
ESI-MS is a soft ionization method for detection and identification of molecules. This mode has the advantages of simultaneous detection of various components and the ability to identify the structure of unknown molecules, and has been widely used in single-cell analysis. Zhang et al. [129] put ESI-MS into practical use for single-cell analysis. They found that when the droplet was generated solely through microfluidic manipulation, the matrix effect could not be ignored in the analysis of single cells due to the consequences of the residual culture medium or metabolic components in the droplet. In order to solve this problem, the authors used a specific extraction solvent for selective microextraction analysis of intracellular components (Fig. 10b). By this method, matrix-free selective detection of intracellular components can be realized. Using this method, author successfully detected glutathione (GSH), adenosine monophosphate (AMP), adenosine diphosphate (ADP), adenosine triphosphate (ATP), L-Glutathione Oxidized (GSSG) and UDP-GlcNAC in single MCF-7 cells. However, this method required microextraction, resulting in a significant increase in the difficulty of operation. In order to solve the operational problems, the probe ESI-MS (PESI-MS) has been proposed for the molecular analysis of cells at the single-cell and subcellular levels. A sharp solid needle probe can be used as an ESI emitter. Gong et al. [130] directly inserted a tungsten probe with a tip diameter of about 1 µum into living cells for enrichment of metabolites, and then detected the enriched molecules through directly ionizing from the tip of the probe. The experimental results show that the method can be applied for the detection of single cell metabolites. Chen et al. [131] also analyzed single-cell lipids using a tungsten probe via PESI-MS to obtain single-cell lipid fingerprints. However, Peretzki et al. [132] recently stumbled upon the fact that electrospray potential can strongly interfere with droplet microfluidic by electrowetting. In order to find a solution, the authors studied different shielding methods to decouple the droplet from the electric field (Fig. 10c). It turned out that it was effective to flood the entire chip into a high voltage.
MALDI-MS is a soft ionization technique, which differs from ESI in that it absorbs laser energy through the matrix and transfers part of the charge to the analyte to ionize it. Xie et al. [133] developed a high-throughput MALDI-MS microfluidic platform for automatic analysis of single-cell phospholipids. The microfluidic chip consisted of microporous array were shown in Fig. 10d. The chip can screen and arrange cells to obtain single cells to be analyzed. Automated high-throughput MALDI-MS imaging analysis of single cells could be performed and the results showed that 8 phospholipids were detected in a single A549 cell, and the strength of phospholipids matched the cell location in MALDI-MS imaging. The device provides a convenient, high-throughput and automated microplatform for the analysis of single cells. By optimizing the size of the micropores, the analysis can be extended to various cell types and different small molecules.
ICP-MS has high sensitivity for trace element analysis of various elements. It has been widely used to detect trace metal or nonmetallic elements which play an important role in cell metabolism and maintenance of normal life activities. In order to study the cell toxicity of CdSe quantum dots (QDs), Yu et al. [134] developed an ICP-MS and microfluidics binding device for the detection of fraction of Cd and Se, which released after cell co-culture with QDs. When the co-cultured cells were injected into the microfluidic channel, the released Cd and Se could be extracted simultaneously through the magnetic solid phase microextraction (MSMPE) column. After optimization of the conditions, the author successfully applied the device to analyze the release of Cd and Se in HepG2 cells incubated with QDs. The experimental results have guided significance for the study of cytotoxicity of CdSe QDs. This method will expand the application prospect for the analysis of cell samples. In addition, Verboket et al. [135] combined the micro-droplets with ICP-MS. Using the incompatibility of oil phase and water phase to generate droplet at the cross channel, it is possible to encapsulate only one cell in each droplet by optimizing the microfluidic manipulation conditions. Due to the influence of oil phase is always not compatible with MS detection, the organic phase was removed through the custom-built transport system of heater and membrane desolvator. Inspired by this study, Wang et al. [136] carried out structural optimization on the basis of the device. An ICP-MS on-line analysis system was designed for droplet-based microchip. The system introduced a microflow nebulizer to avoid plasma instability caused by a large number of organic solvents. The device is simple in structure and easy to be operated and presents a good application prospect in the analysis of trace elements in single cells.
In addition, the special design based on microfluidic can also bring unexpected convenience for mass spectrometric detection. Flexible use of the characteristics of microfluidic chips can achieve faster, more accurate and concise biological analysis. In order to obtain more accurate information for the analysis of single cell components, the steps of separation and purification are very important. The separation process directly affects the accuracy of the results. Huang et al. [124] designed a microfluidic method based on in situ single cell recognition system for single cell extraction. The system was shown to be able to isolate an entire cell and remove interference from the solution components. Same for in situ separation, a helical channel microfluidic chip is used to generate Dean flow for cell separation and MS detection. The system can effectively reduce cell aggregation and significantly improve the efficiency of single-cell MS analysis [125]. In order to improve the analysis efficiency of microfluidic chip-MS, the design of multi-channel microfluidic chip is particularly important. The introduction of multi-channel microfluidic chip can provide the possibility of multi-channel parallel work, thus greatly reducing the time of sample pretreatment and analysis. Jie et al. [126] reviewed multi-channel microfluidic chips and pointed out their importance.
MS has shown great power in the analysis of single cells, but its application is not limited to single cells. Metabolite detection is also very important and can provide us with supplementary information for cell detection. The presence of metabolites in complex microenvironments at very low levels poses challenges to analysis, but MS also plays an important role in this area due to its advantages. At present, the studies related to metabolism mainly focus on the metabolites of tissues and organs, as well as the evaluation of drug toxicity. Such as Mao et al. [137] developed an integrated microfluidic device-MS system for simulating drug metabolism in the liver and HepG2 cell toxicity. Zheng et al. [14] simulated drug metabolism in a cell co-culture microcapsule model. Briefly, in this study, a 3D tumor-endothelial co-culture model was constructed in a microfluidics channel, and drug metabolites were measured by MS to determine drug resistance during simulated tumor treatment.
3.5. Electrochemical detectionElectrochemistry technology is one of the most commonly methods used in the field of analysis. It has the advantages of high sensitivity and fast response, and is widely used in biological research to record nerve signals and detect biomolecules offering new insights into physiology and pathology [138-141]. In recent years, with the maturity of micromanufacturing technology, the integrated microelectrodes on microfluidic platforms for detecting biomolecules have been gradually popularized. Lin et al. [142] developed a method of microchip electrophoresis based on tunable aptamer for multiplex protein assay. Proteins were separated by modifying with different lengths of aptamers. Platelet-derived growth factor B-chain, thrombin and human vascular endothelial growth factor 165 were quantitatively assayed on this chip with a good dynamic range and satisfactory relative standard deviation values. This kind of electrochemical platform also was applied in detecting the metabolites and exosomes of single cells to study the physiological function and stress response of cells. Senel et al. [143] assembled three electrodes on a microfluidic chip to quickly detect dopamine (DA) which was an important biomolecule that regulated the nervous system (Fig. 11a). With its high sensitivity, rapid response and low detection limit for DA, the device provided an important diagnostic method for a variety of DA-deficient neurological diseases. However, microelectrode alone is not adequate for detecting other biomarkers with low sensitivity and low content. Detection of these substances requires the signals to be converted or amplified. In Fig. 11b, Safaei et al. [144] reported a microfluidic chip for detecting CTC. The electrochemical analysis chip combined with an enzyme-linked immunosorbent assay (ELISA) method can be used to detect lower concentrations of cancer cells in whole blood. In this study, a signal transformation method was used to detect cancer cells. As a signal source, the redox reporter was conjugated with the cancer cells to form complexes through antigen and antibody specific recognition. In addition, preconcentration is also a method to increase the detection sensitivity. Methylated DNA is a biomarker for early diagnosis of cancer, and its detection is of great clinical significance. However, its scarcity makes detection to be a great challenge. Hong et al. [145] used ion concentration polarization to preconcentrate methylated DNA and performed in situ electrochemical detection on a chip. Moreover, Feng et al. [146] demonstrated a microfluidic chip that enhanced the detection signal via photoelectrochemistry (PEC). The mechanism was shown in Fig. 11c. O2 on the cathode consumed electrons and generated superoxide anions radical (˙O2−) which reacted with co-catalyst generating O2 and H2O2 and amplifying signals. H2O2 is decomposed to be O2. The increased O2 further consumed electrons and promoted the separation of electron-hole pairs, thus effectively increasing the PEC effect. The results showed that the device exhibited good linearity in the concentration range of 0.1-100 pg/mL. This study provided a new signal amplification strategy for the detection of biomarkers and provided a new promising detection method for clinical medicine and disease diagnosis.
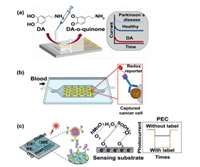
|
Download:
|
| Fig. 11. Electrochemical detection on microfluidic chips. (a) Three electrodes were modified on a microfluidic chip for the quick detection of DA. Copied with permission [143]. Copyright 2020, American Chemical Society. (b) The electrochemical analysis chip combined with an ELISA. Copied with permission [144]. Copyright 2015, American Chemical Society. (c) PEC analysis of the cells. WE: working microelectrode; RE: reference microelectrode; CE: counter microelectrode. Copied with permission [146]. Copyright 2021, the Royal Society of Chemistry. | |
Microfluidic impedance cytometry is a label-free and high-throughput single-cell analysis method that stratifies the heterogeneity of cellular systems based on electrophysiology properties and can be used to detect, count and analyze cells [147]. With breakthroughs in research, microfluidic impedance cell technology has been widely used in medical diagnosis, drug evaluation and precision medicine. Evander et al. [148] demonstrated a platelet analysis platform based on a microfluidic impedance cytometer. David et al. [149] developed a microfluidic single-cell impedance cytometer that performed a WBC differential count and analysis. Results from patient samples were similar to those from commercial blood analysis devices, indicating potential commercial and clinical value of the device. Lin et al. [150] recently realized the electrical detection of cancer cell surface proteins using multifrequency microfluidic impedance cytometry, providing a potential solution for studying the expression of target antigens on the surface of cancer cells. This approach was not limited to the analysis of cancer cells, but also applied to a wide range of markers and cell types. Moreover, the introduction of coded biomarkers to identify objects is also an analytical approach. A hydrogel bead was introduced for electroquantification of protein targets to perform versatile and multiplexed detection of biomolecules by means of unique impedance signatures generated during coulter counting [151]. In a nutshell, with the diversification of microchips, more and more novel chip designs provided more possibilities and opportunities for experiments. Honrado et al. [152] reviewed the recent microfluidic impedance cytometry analysis of multi-parameter cell characterization and subgroup feature differentiation in detail, including design, analysis and application.
4. ConclusionThe methods and principles of microfluidics for cell screening, separating, purifying, capturing and analyzing are reviewed in this paper. The means involved are varied, and the ways of capture and analysis based on different principles exhibit their own advantages and limitations, so it is necessary to design the most appropriate methods according to the physical and chemical characteristics of the target cells. With the support of high integration, high sensitivity and high throughput, microfluidic system has highlighted great advantages in the analysis of cell nucleic acid, protein, exosome and other cytokines, which made important contributions to clinical medicine such as early detection of diseases and drug tolerance. But that is not the end of the story. We need to go further than that. The future research direction should focus on improving the operational ability of cells and more sensitive analysis methods, and gradually developing single-cell analysis at subcellular or even single-molecule level, in order to promote a deeper, more accurate and more comprehensive understanding of the various functions and mechanisms of cytokines in regulating physiological processes. We believe that in the near future, microfluidic-based cell analysis will lead us to explore previously inaccessible levels, allow us to have a new understanding of the units that make up ourselves, and drive the development of the medical field.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentsThis work was supported by the National Natural Science Foundation of China (No. 21874120) and the Fundamental Research Funds for the Central Universities (No. 2652019112, 2652018004) and the open fund of Key Laboratory of Optic-electric Sensing and Analytical Chemistry for Life Science, MOE, Qingdao University of Science and Technology (No. OESACLS202004).
| [1] |
Q. Huang, S. Mao, M. Khan, M. J.-Lin, Analyst 144 (2019) 808-823. DOI:10.1039/C8AN01079J |
| [2] |
R. Zenobi, Analytical and Biological Perspectives 342 (2013) 1243259. |
| [3] |
F.C. Tang, K.Q. Lao, M.A. Surani, Nat. Methods 8 (2011) S6-S11. DOI:10.1038/nmeth.1557 |
| [4] |
D.A. Lawson, N.R. Bhakta, K. Kessenbrock, et al., Nature 526 (2015) 131-135. DOI:10.1038/nature15260 |
| [5] |
F.C. Tang, C. Barbacioru, Y.Z. Wang, et al., Nat. Methods 6 (2009) 377-382. DOI:10.1038/nmeth.1315 |
| [6] |
Y. Kieffer, H.R. Hocine, G. Gentric, et al., Cancer Discov 10 (2020) 1330-1351. DOI:10.1158/2159-8290.CD-19-1384 |
| [7] |
A.C. Habermann, A.J. Gutierrez, L.T. Bui, et al., Sci. Adv. 6 (2020) 15. |
| [8] |
H. Mathys, J. Davila-Velderrain, Z. Peng, et al., Nature 570 (2019) 332-337. DOI:10.1038/s41586-019-1195-2 |
| [9] |
T. Stuart, A. Butler, P. Hoffman, et al., Cell 177 (2019) 1888-1902. DOI:10.1016/j.cell.2019.05.031 |
| [10] |
T. Masuda, R. Sankowski, O. Staszewski, et al., Nature 566 (2019) 388-392. DOI:10.1038/s41586-019-0924-x |
| [11] |
S. Lindstrom, H. Andersson-Svahn, Lab Chip 10 (2010) 3363-3372. DOI:10.1039/c0lc00150c |
| [12] |
Q. Zhang, S. Feng, L. Lin, S. Mao, M. J.-Lin, Chem. Soc. Rev. 50 (2021) 5333-5348. DOI:10.1039/D0CS01516D |
| [13] |
Y. Zheng, Z. Wu, M. Khan, et al., Anal. Chem. 91 (2019) 12283-12289. DOI:10.1021/acs.analchem.9b02434 |
| [14] |
Y. Zheng, Z. Wu, M. J.-Lin, L. Lin, Chin. Chem. Lett. 31 (2020) 451-454. DOI:10.1016/j.cclet.2019.07.036 |
| [15] |
S. Feng, S. Mao, Q. Zhang, et al., ACS Sensors 4 (2019) 521-527. DOI:10.1021/acssensors.8b01696 |
| [16] |
W. Li, M. Khan, S. Mao, et al., J. Pharm. Anal. 8 (2018) 210-218. DOI:10.1016/j.jpha.2018.07.005 |
| [17] |
S. Mao, Q. Zhang, H. Li, et al., Chem. Sci. 9 (2018) 7694-7699. DOI:10.1039/C8SC03027H |
| [18] |
S. Mao, Q. Zhang, H. Li, et al., Anal. Chem. 90 (2018) 9637-9643. DOI:10.1021/acs.analchem.8b02653 |
| [19] |
S. Mao, W. Zhang, Q. Huang, et al., Angew. Chem. Int. Edit. 57 (2018) 236-240. DOI:10.1002/anie.201710273 |
| [20] |
J. Dou, S. Mao, H. Li, M. J-Lin, Anal. Chem. 92 (2020) 892-898. DOI:10.1021/acs.analchem.9b03681 |
| [21] |
T. Xie, N. Li, S. Mao, et al., Cell Heterogeneity Revealed by On-Chip Angiogenic Endothelial Cell Migration 5 (2020) 3857-3862. |
| [22] |
C. Alix-Panabieres, K. Pantel, Clin. Chem. 59 (2013) 110-118. DOI:10.1373/clinchem.2012.194258 |
| [23] |
H. Jeon, B. Jundi, K. Choi, et al., Lab Chip 20 (2020) 3612-3624. DOI:10.1039/D0LC00675K |
| [24] |
Y. Huang, Y. Sheng, S. Chao, et al., Lab Chip 20 (2020) 4342-4348. DOI:10.1039/D0LC00895H |
| [25] |
J. Zhou, I. Papautsky, Lab Chip 19 (2019) 3416-3426. DOI:10.1039/C9LC00786E |
| [26] |
V.J. Lyons, A. Helms, D. Pappas, Anal. Chim. Acta 1076 (2019) 154-161. DOI:10.1016/j.aca.2019.05.040 |
| [27] |
K.C. Andree, A. Mentink, A.T. Nguyen, et al., Lab Chip 19 (2019) 1006-1012. DOI:10.1039/C8LC01158C |
| [28] |
S.L. Stott, C.H. Hsu, D.I. Tsukrov, et al., Proc. Natl. Acad. Sci. U. S. A. 107 (2010) 18392-18397. |
| [29] |
S. Nagrath, L.V. Sequist, S. Maheswaran, et al., Nature 450 (2007) 1235-1239. DOI:10.1038/nature06385 |
| [30] |
H. Pei, L. Li, Z. Han, et al., Lab Chip 20 (2020) 3854-3875. DOI:10.1039/D0LC00577K |
| [31] |
S.M. Santana, H. H. N Liu, et al., Biomed. Microdevices 14 (2012) 401-407. DOI:10.1007/s10544-011-9616-5 |
| [32] |
Y. D.-Oh, J. Y.-Bang, Nat. Rev. Clin. Oncol. 17 (2020) 33-48. DOI:10.1038/s41571-019-0268-3 |
| [33] |
C. Spiess, Q. Zhai, P.J. Carter, Mol. Immunol. 67 (2015) 95-106. DOI:10.1016/j.molimm.2015.01.003 |
| [34] |
Y. Song, Z. Zhu, Y. An, et al., Anal. Chem. 85 (2013) 4141-4149. DOI:10.1021/ac400366b |
| [35] |
M. Mashreghi, P. Zamani, S.A. Moosavian, M.R. Jaafari, Nanoscale Res. Lett. 15 (2020) 101. DOI:10.1186/s11671-020-03334-9 |
| [36] |
T. Y.-Kang, Y.J. Kim, J. Bu, et al., Sens. Actuators B-Chem. 260 (2018) 320-330. DOI:10.1016/j.snb.2017.12.157 |
| [37] |
T. Wang, C.Y. L. M Chen, et al., Biotechnol. Adv. 37 (2019) 28-50. DOI:10.1016/j.biotechadv.2018.11.001 |
| [38] |
P. Rothlisberger, M. Hollenstein, Adv. Drug Deliv. Rev. 134 (2018) 3-21. DOI:10.1016/j.addr.2018.04.007 |
| [39] |
M.R. Dunn, R.M. Jimenez, J.C. Chaput, Nat. Rev. Chem. 1 (2017) 16. DOI:10.1038/s41570-017-0016 |
| [40] |
Q. Chen, J. Wu, Y. Zhang, Z. Lin, M. J.-Lin, Lab Chip 12 (2012) 5180-5185. DOI:10.1039/c2lc40858a |
| [41] |
M. Lin, F. J.-Chen, T. Y.-Lu, et al., Accounts Chem. Res. 47 (2014) 2941-2950. DOI:10.1021/ar5001617 |
| [42] |
L. Zhao, C. Tang, L. Xu, et al., Small 12 (2016) 1072-1081. DOI:10.1002/smll.201503188 |
| [43] |
C.S. Lin, Y.C. Tsai, K.F. Hsu, G.B. Lee, Lab Chip 21 (2021) 725-734. DOI:10.1039/D0LC01333A |
| [44] |
M. Hejazian, W. Li, N. Nam-Trung, Lab Chip 15 (2015) 959-970. DOI:10.1039/C4LC01422G |
| [45] |
W. Shi, S. Wang, A. Maarouf, et al., Lab Chip 17 (2017) 3291-3299. DOI:10.1039/C7LC00333A |
| [46] |
F. Zhang, L.L. Wu, W.D. Nie, et al., Anal. Chem. 91 (2019) 15726-15731. DOI:10.1021/acs.analchem.9b03920 |
| [47] |
W. Zhao, R. Cheng, B.D. Jenkins, et al., Lab Chip 17 (2017) 3097-3111. DOI:10.1039/C7LC00680B |
| [48] |
K. Xiong, W. Wei, Y.J. Jin, et al., Adv. Mater. 28 (2016) 7929-7935. DOI:10.1002/adma.201601643 |
| [49] |
P. Liu, P. Jonkheijm, L. Terstappen, M. Stevens, Cancers 12 (2020) 26. |
| [50] |
L.A. Luo, Y.Q. He, Cancer Med 9 (2020) 4207-4231. DOI:10.1002/cam4.3077 |
| [51] |
Y. Sui, C.J. Teo, P.S. Lee, Y.T. Chew, C. Shu, Int. J. Heat Mass Transf. 53 (2010) 2760-2772. DOI:10.1016/j.ijheatmasstransfer.2010.02.022 |
| [52] |
M.E. Warkiani, G. Guan, K.B. Luan, et al., Lab Chip 14 (2014) 128-137. DOI:10.1039/C3LC50617G |
| [53] |
A. Shiriny, M. Bayareh, Chem. Eng. Sci. 229 (2021) 116102. DOI:10.1016/j.ces.2020.116102 |
| [54] |
H. Tavassoli, P. Rorimpandey, Y.C. Kang, et al., Small 17 (2021) 2006176. DOI:10.1002/smll.202006176 |
| [55] |
Q. Zhao, D. Yuan, J. Zhang, W. Li, Micromachines 11 (2020) 461. DOI:10.3390/mi11050461 |
| [56] |
H. P.-Tsou, H. P.-Chiang, T. Z.-Lin, et al., Lab Chip 20 (2020) 4007-4015. DOI:10.1039/D0LC00663G |
| [57] |
E. Guzniczak, O. Otto, G. Whyte, et al., Lab Chip 20 (2020) 614-625. DOI:10.1039/C9LC01000A |
| [58] |
R.K. Gao, L. Cheng, S.Y. Wang, et al., Talanta 207 (2020) 8. |
| [59] |
S. Karimi, P. Mehrdel, J. Farre-Llados, J. Casals-Terre, Lab Chip 19 (2019) 3249-3260. DOI:10.1039/C9LC00690G |
| [60] |
P. Dettinger, W. Wang, N. Ahmed, et al., Lab Chip 20 (2020) 4246-4254. DOI:10.1039/D0LC00687Dyz%$https://www.cnki.com.cn/Article/CJFDTOTAL-GCLX201101034.htm |
| [61] |
H. Pei, L. Li, Y. Wang, et al., Anal. Chem. 91 (2019) 11078-11084. DOI:10.1021/acs.analchem.9b01647 |
| [62] |
Z. Wu, H. Jiang, L. Zhang, et al., Lab Chip 19 (2019) 3922-3930. DOI:10.1039/C9LC00874H |
| [63] |
C. Richard, A. Fakhfouri, M. Colditz, et al., Lab Chip 19 (2019) 4043-4051. DOI:10.1039/C9LC00804G |
| [64] |
C. Devendran, K. Choi, J. Han, et al., Lab Chip 20 (2020) 2674-2688. DOI:10.1039/D0LC00397B |
| [65] |
N. Zhang, J.P. Zuniga-Hertz, E.Y. Zhang, et al., Lab Chip 21 (2021) 904-915. DOI:10.1039/D0LC01012J |
| [66] |
E. Brouzes, M. Medkova, N. Savenelli, et al., Proc. Natl. Acad. Sci. U. S. A. 106 (2009) 14195-14200. DOI:10.1073/pnas.0903542106 |
| [67] |
S.Y. Teh, R. Lin, L.H. Hung, A.P. Lee, Lab Chip 8 (2008) 198-220. DOI:10.1039/b715524g |
| [68] |
A.B. Theberge, F. Courtois, Y. Schaerli, et al., Angew. Chem. Int. Edit. 49 (2010) 5846-5868. DOI:10.1002/anie.200906653 |
| [69] |
F. Del Ben, M. Turetta, G. Celetti, et al., Angew. Chem. Int. Edit. 55 (2016) 8581-8584. DOI:10.1002/anie.201602328 |
| [70] |
G. Leonaviciene, K. Leonavicius, R. Meskys, L. Mazutis, Lab Chip 20 (2020) 4052-4062. DOI:10.1039/D0LC00660B |
| [71] |
M. Wang, M.H. Nai, J. R.Y.-Huang, et al., Lab Chip 21 (2021) 764-774. DOI:10.1039/D0LC01294G |
| [72] |
R. Zilionis, J. Nainys, A. Veres, et al., Nat. Protoc. 12 (2017) 44-73. DOI:10.1038/nprot.2016.154 |
| [73] |
C.G. Yang, R.Y. Pan, Z.R. Xu, Chin. Chem. Lett. 26 (2015) 1450-1454. DOI:10.1016/j.cclet.2015.10.016 |
| [74] |
W. Zhang, N. Li, L. Lin, et al., Small 16 (2020) 1903402. DOI:10.1002/smll.201903402 |
| [75] |
M. Jahangiri, S. Khosravi, H. Moghtaderi, et al., Sens. Actuators B-Chem. 297 (2019) 126733. DOI:10.1016/j.snb.2019.126733 |
| [76] |
B. Chen, W. Le, Y. Wang, et al., Theranostics 6 (2016) 1887-1898. DOI:10.7150/thno.16358 |
| [77] |
I. S.-Han, C. Huang, A. Han, Lab Chip 20 (2020) 3832-3841. DOI:10.1039/D0LC00710B |
| [78] |
C. Y.-Kung, K.R. Niazi, Y. P.-Chiou, Lab Chip 21 (2021) 1049-1060. DOI:10.1039/D0LC00853B |
| [79] |
D. Yang, Y. Ai, Lab Chip 19 (2019) 3609-3617. DOI:10.1039/C9LC00819E |
| [80] |
X. Huang, K. Torres-Castro, W. Varhue, et al., Lab Chip 21 (2021) 835-843. DOI:10.1039/D0LC01211D |
| [81] |
H. K.-Han, I. S.-Han, A.B. Frazier, Lab Chip 9 (2009) 2958-2964. DOI:10.1039/b909753h |
| [82] |
I. S.-Han, M. S.-Lee, D. Y.-Joo, H. K.-Han, Lab Chip 11 (2011) 3864-3872. DOI:10.1039/c1lc20413k |
| [83] |
I. S.-Han, H.S. Kim, H. K.-Han, A. Han, Lab Chip 19 (2019) 4128-4138. DOI:10.1039/C9LC00850K |
| [84] |
L. Lin, S. Y.-Chu, J.P. Thiery, C.T. Lim, I. Rodriguez, Lab Chip 13 (2013) 714-721. DOI:10.1039/c2lc41070b |
| [85] |
D. Di Carlo, N. Aghdam, L.P. Lee, Anal. Chem. 78 (2006) 4925-4930. DOI:10.1021/ac060541s |
| [86] |
Q. Chen, J. Wu, Y. Zhang, Z. Lin, M. J.-Lin, Lab Chip 12 (2012) 5180-5185. DOI:10.1039/c2lc40858a |
| [87] |
X. Qin, S. Park, S.P. Duffy, et al., Lab Chip 15 (2015) 2278-2286. DOI:10.1039/C5LC00226E |
| [88] |
Y. Liu, T. Li, M. Xu, et al., Lab Chip 19 (2019) 68-78. DOI:10.1039/C8LC01048J |
| [89] |
J. Ko, N. Bhagwat, S.S. Yee, et al., Lab Chip 17 (2017) 3086-3096. DOI:10.1039/C7LC00703E |
| [90] |
D. Yuan, Q. Zhao, S. Yan, et al., Lab Chip 19 (2019) 2811-2821. DOI:10.1039/C9LC00482C |
| [91] |
Z. Ma, Y. Zhou, D.J. Collins, Y. Ai, Lab Chip 17 (2017) 3176-3185. DOI:10.1039/C7LC00678K |
| [92] |
Y. Song, H. Yin, W.E. Huang, Curr. Opin. Chem. Biol. 33 (2016) 1-8. |
| [93] |
Y. Lyu, X. Yuan, A. Glidle, et al., Lab Chip 20 (2020) 4235-4245. DOI:10.1039/D0LC00679C |
| [94] |
X. Hu, D. Zhu, M. Chen, et al., Lab Chip 19 (2019) 2549-2556. DOI:10.1039/C9LC00361D |
| [95] |
M. Khan, S. Mao, W. Li, M. J.-Lin, Chem.-Eur. J. 24 (2018) 15398-15420. DOI:10.1002/chem.201800305 |
| [96] |
P. Ramachandran, R. R. J Dobie, et al., Nature 575 (2019) 512-518. DOI:10.1038/s41586-019-1631-3 |
| [97] |
M.Y. Batiuk, A. Martirosyan, J. Wahis, et al., Nat. Commun. 11 (2020) 1220. DOI:10.1038/s41467-019-14198-8 |
| [98] |
I. Korsunsky, N. Millard, J. Fan, et al., Nat. Methods 16 (2019) 1289-1296. DOI:10.1038/s41592-019-0619-0 |
| [99] |
Y. Fan, D.Q. Dong, et al., Lab Chip 18 (2018) 1151-1173. DOI:10.1039/C7LC01333G |
| [100] |
N. Li, W. Zhang, Y. Li, M. J.-Lin, TrAC-Trends Anal. Chem. 117 (2019) 200-214. DOI:10.1016/j.trac.2019.05.029 |
| [101] |
Q. Zhang, S. Feng, W. Li, et al., Angew. Chem. Int. Edit. 60 (2021) 8483-8487. DOI:10.1002/anie.202016171 |
| [102] |
H. Guo, X. Song, W. Lei, et al., Angew. Chem. Int. Edit. 58 (2019) 12195-12199. DOI:10.1002/anie.201907605 |
| [103] |
M.E. Gallina, T.J. Kim, M. Shelor, et al., Anal. Chem. 89 (2017) 6472-6481. DOI:10.1021/acs.analchem.7b00414 |
| [104] |
E.X. Ng, M.A. Miller, T.Y. Jing, C.H. Chen, Biosens. Bioelectron. 81 (2016) 408-414. DOI:10.1016/j.bios.2016.03.002 |
| [105] |
J. Liu, G. Sun, C. S.-Wei, et al., Lab Chip 20 (2020) 1939-1946. DOI:10.1039/C9LC01226E |
| [106] |
Y. Wang, Q. Li, H. Shi, et al., Lab Chip 20 (2020) 4632-4637. DOI:10.1039/D0LC00677G |
| [107] |
C.H. Camp Jr., S. Yegnanarayanan, A.A. Eftekhar, H. Sridhar, A. Adibi, Opt. Express 17 (2009) 22879-22889. DOI:10.1364/OE.17.022879 |
| [108] |
C. Zhang, C. K.-Huang, B. Rajwa, et al., Optica 4 (2017) 103-109. DOI:10.1364/OPTICA.4.000103 |
| [109] |
Y. Wang, Q. Zhang, W. Yuan, et al., Lab Chip 21 (2021) 196-204. DOI:10.1039/D0LC01006E |
| [110] |
D.M.D. Siu, K.C.M. Lee, M.C.K. Lo, et al., Lab Chip 20 (2020) 3696-3708. DOI:10.1039/D0LC00542H |
| [111] |
A. Kleiber, A. Ramoji, G. Mayer, et al., Lab Chip 20 (2020) 1676-1686. DOI:10.1039/D0LC00244E |
| [112] |
Y. Luo, J. Yang, X. Zheng, et al., Lab Chip 21 (2021) 75-82. DOI:10.1039/D0LC00917B |
| [113] |
P. Zhang, W. Wang, H. Fu, et al., Lab Chip 20 (2020) 4466-4473. DOI:10.1039/D0LC00538J |
| [114] |
K. Mutafopulos, P.J. Lu, R. Garry, P. Spink, D.A. Weitz, Lab Chip 20 (2020) 3914-3921. DOI:10.1039/D0LC00723D |
| [115] |
N. Hamada, Y. Hashi, S. Yamaki, et al., Chin. Chem. Lett. 30 (2019) 99-102. DOI:10.1016/j.cclet.2018.10.029 |
| [116] |
W. Zhang, Q. Zhang, J.-M. Lin, Cell Analysis on Microfluidics Combined with Mass Spectrometry[J]. Anal. Sci. 37 (2021) 249-260. DOI:10.2116/analsci.20R006 |
| [117] |
S. Mao, W. Li, Q. Zhang, et al., TrAC-Trends Anal. Chem. 107 (2018) 43-59. DOI:10.1016/j.trac.2018.06.019 |
| [118] |
L. Yin, Z. Zhang, Y. Liu, Y. Gao, J. Gu, Analyst 144 (2019) 824-845. DOI:10.1039/C8AN01190G |
| [119] |
S. Theiner, K. Loehr, G. Koellensperger, L. Mueller, N. Jakubowski, J. Anal. At. Spectrom. 35 (2020) 1784-1813. DOI:10.1039/D0JA00194E |
| [120] |
Y.Y. Yang, Y.Y. Huang, J.H. Wu, et al., TrAC-Trends Anal. Chem. 90 (2017) 14-26. DOI:10.1016/j.trac.2017.02.009 |
| [121] |
B. Domon, R. Aebersold, Science 312 (2006) 212-217. DOI:10.1126/science.1124619 |
| [122] |
L. Nie, G.B. Xu, X.Y. Wang, et al., Chin. Chem. Lett. 24 (2013) 491-493. DOI:10.1016/j.cclet.2013.03.053 |
| [123] |
D. Gao, H. Liu, Y. Jiang, M. J.-Lin, Lab Chip 13 (2013) 3309-3322. DOI:10.1039/c3lc50449b |
| [124] |
Q. Huang, S. Mao, M. Khan, et al., Chem. Sci. 11 (2020) 253-256. DOI:10.1039/C9SC05143K |
| [125] |
Q. Huang, S. Mao, M. Khan, et al., Chem. Commun. 54 (2018) 2595-2598. DOI:10.1039/C7CC09608A |
| [126] |
M.S. Jie, S.F. Mao, H.F. Li, J.M. Lin, Chin. Chem. Lett. 28 (2017) 1625-1630. DOI:10.1016/j.cclet.2017.05.024 |
| [127] |
N. Xu, H.F. Lin, S. Lin, et al., Anal. Chem. 93 (2021) 2273-2280. DOI:10.1021/acs.analchem.0c04147 |
| [128] |
R.D. Pedde, H. Li, C.H. Borchers, M. Akbari, Trends Biotechnol 35 (2017) 954-970. DOI:10.1016/j.tibtech.2017.06.006 |
| [129] |
C. X.-Zhang, W. Z.-Wei, Y. X.-Gong, et al., Sci Rep 6 (2016) 24730. DOI:10.1038/srep24730 |
| [130] |
X. Gong, Y. Zhao, S. Cai, et al., Anal. Chem. 86 (2014) 3809-3816. DOI:10.1021/ac500882e |
| [131] |
F. Chen, L. Lin, J. Zhang, et al., Anal. Chem. 88 (2016) 4354-4360. DOI:10.1021/acs.analchem.5b04749 |
| [132] |
A.J. Peretzki, S. Schmidt, E. Flachowsky, et al., Lab Chip 20 (2020) 4456-4465. DOI:10.1039/D0LC00936A |
| [133] |
W. Xie, D. Gao, F. Jin, et al., Anal. Chem. 87 (2015) 7052-7059. DOI:10.1021/acs.analchem.5b00010 |
| [134] |
X. Yu, B. Chen, M. He, H. Wang, B. Hu, Talanta 179 (2018) 279-284. DOI:10.1016/j.talanta.2017.11.013 |
| [135] |
P.E. Verboket, O. Borovinskaya, N. Meyer, et al., Anal. Chem. 86 (2014) 6012-6018. DOI:10.1021/ac501149a |
| [136] |
H. Wang, B. Chen, M. He, B. Hu, Anal. Chem. 89 (2017) 4931-4938. DOI:10.1021/acs.analchem.7b00134 |
| [137] |
S. Mao, D. Gao, W. Liu, et al., Lab Chip 12 (2012) 219-226. DOI:10.1039/C1LC20678H |
| [138] |
K. Wang, C.L. Frewin, D. Esrafilzadeh, et al., Adv. Mater. 31 (2019) 1805867. DOI:10.1002/adma.201805867 |
| [139] |
V. Mani, B. Devadas, M. S.-Chen, Biosens. Bioelectron. 41 (2013) 309-315. DOI:10.1016/j.bios.2012.08.045 |
| [140] |
J. Ping, J. Wu, Y. Wang, Y. Ying, Biosens. Bioelectron. 34 (2012) 70-76. DOI:10.1016/j.bios.2012.01.016 |
| [141] |
N. Gupta, V. Renugopalakrishnan, D. Liepmann, R. Paulmurugan, B.D. Malhotra, Biosens. Bioelectron. 141 (2019) 111435. DOI:10.1016/j.bios.2019.111435 |
| [142] |
X. Lin, Q. Chen, W. Liu, et al., Biosens. Bioelectron. 63 (2015) 105-111. DOI:10.1016/j.bios.2014.07.013 |
| [143] |
M. Senel, E. Dervisevic, S. Alhassen, et al., Anal. Chem. 92 (2020) 12347-12355. DOI:10.1021/acs.analchem.0c02032 |
| [144] |
T.S. Safaei, R.M. Mohamadi, E.H. Sargent, S.O. Kelley, ACS Appl. Mater. Interfaces 7 (2015) 14165-14169. DOI:10.1021/acsami.5b02404 |
| [145] |
S.A. Hong, J. Y.-Kim, S.J. Kim, S. Yang, Biosens. Bioelectron. 107 (2018) 103-110. DOI:10.1016/j.bios.2018.01.067 |
| [146] |
J. Feng, T. Wu, Q. Cheng, et al., Lab Chip 21 (2021) 378-384. DOI:10.1039/D0LC01063D |
| [147] |
K.C. Cheung, M. Di Berardino, G. Schade-Kampmann, et al., Cytometry Part A 77A (2010) 648-666. DOI:10.1002/cyto.a.20910 |
| [148] |
M. Evander, A.J. Ricco, J. Morser, et al., Lab Chip 13 (2013) 722-729. DOI:10.1039/c2lc40896a |
| [149] |
D. Holmes, D. Pettigrew, C.H. Reccius, et al., Lab Chip 9 (2009) 2881-2889. DOI:10.1039/b910053a |
| [150] |
Z. Lin, Y. S.-Lin, P. Xie, et al., Sci Rep 10 (2020) 3015. DOI:10.1038/s41598-020-57540-7 |
| [151] |
T.W. Cowell, E. Valera, A. Jankelow, et al., Lab Chip 20 (2020) 2274-2283. DOI:10.1039/D0LC00243G |
| [152] |
C. Honrado, P. Bisegna, N.S. Swami, F. Caselli, Lab Chip 21 (2021) 22-54. DOI:10.1039/D0LC00840K |
 2022, Vol. 33
2022, Vol. 33 

