b Department of Chemistry and COSDAF (Centre of Super-Diamond and Advanced Films), City University of Hong Kong, Hong Kong, China;
c Key Laboratory of Biochip Technology, Biotech and Health Centre, Shenzhen Research Institute of City University of Hong Kong, Shenzhen 518057, China;
d College of Food Engineering, Qingdao Institute of Technology, Qingdao 266300, China;
e Department of Chemistry, The Chinese University of Hong Kong, Hong Kong, China;
f Department of Chemistry, National University of Singapore, Singapore 117543, Singapore
Microarray technology allows thousands of biological samples to be screened on a planar surface rapidly and simultaneously. It features a number of prominent advantages, such as parallelization, miniaturization and automation. Through the development of peptides [1], proteins and so on, microarray strategy has been widely utilized to anchor DNAs [2], small molecules [3], peptides [4, 5] and proteins [6-10] for many biological applications including protein functional annotation, inhibitor development, biomarker discovery and disease diagnosis [11, 12].
The success of microarray fabrication process heavily depends on biomolecule immobilization. Researchers have spent tremendous efforts in developing various efficient immobilization approaches for microarray fabrication. The immobilization strategies can be broadly classified into two major types in terms of noncovalent and covalent immobilization. Seminal examples of noncovalent immobilization methods include the polyhistidine-nickel nitrilotriacetate acid complex and the biotin-avidin interactions [8, 13]. On the other hand, numerous chemoselective reactions have been developed for covalent microarray immobilization such as click chemistry, thiol-ene chemistry, Diels-Alder reaction and Staudinger ligation [14-17, 21]. Recently we have also developed two robust immobilization methods based on biocompatible reactions, i.e., cysteine/2-cyanobenzothiazole [18], and trans-cyclooctene/tetrazine [19, 20]. The first approach provides a convenient tool for immobilization of peptides with N-terminal cysteine residue. The latter immobilization method is extremely fast, and it can immobilize proteins within a short time.
Despite these advancements, there is still a lack of reversible immobilization methods, which can dramatically save the time and costs during the microarray fabrication process [6]. This creates an urgent need to develop novel reversible immobilization methods for further expansion of the microarray field. Recently, Trilla et al. [21] has introduced an elegant and reversible immobilization method based on the interaction between biarsenical dye and tetracysteine motif. The method allows direct detection of the immobilization process by monitoring fluorescence enhancement of the biarsenical dye. It can also be used to immobilize proteins directly from cellular lysates. Nevertheless, the method requires multi-step synthesis of biarsenic dyes, which might restrict its wide usage in biomedical applications.
In this study, we developed a new and facile reversible immobilization method based on the reaction between thiol and quinone. Our method takes advantage of a commercially available molecule named L-3, 4-dihydroxyphenylalanine (L-Dopa). L-Dopa has been found in mussel food proteins (Mfps), and it makes Mfps wet adhesive by forming various physical and chemical crosslinking with solid surface [22, 23]. Due to its unique property, L-Dopa has been widely used to design materials for different applications including biosensing, drug delivery and biomedical adhesives [24, 25]. In addition, L-dopamine can be oxidized into reactive quinone species, which might induce neurotoxicity and cause Parkinson's disease [26-29].
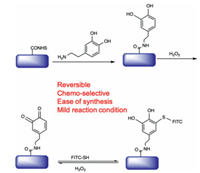
In our approach, we first fabricated dopamine-functionalized slides through standard procedure (Scheme 1). A quinone-functionalized slide was subsequently obtained through H2O2 oxidation process. Various thiol-containing molecules can be linked to the quinone-functionalized slides via thioether linker, which could be cleaved under H2O2 treatment to regenerate the quinone group on the slide surface, thus enabling microarray immobilization in a reversible manner.
|
Download:
|
| Scheme 1. Schematic representation of the reversible microarray immobilization approach based on thiol and quinone reaction. | |
To establish the method, we first carried out detailed HPLC study on the model reaction between glutathione (GSH) and quinone. Hydrogen peroxide was added to a solution of L-Dopa in PBS buffer (pH 7.4), and the reaction was analysed by HPLC and MS. As shown in Fig. 1, the peak of L-Dopa decreased when H2O2 reacted with L-Dopa. A new peak corresponding to quinone product appeared (Fig. 1B and Fig. S4 in Supporting information). Subsequently GSH was added to the quinone solution. The quinone peak disappeared while a new peak with retention time of 11 min appeared (Fig. 1C and Fig. S6 in Supporting information). The new peak corresponds to the quinone-thiol adduct (GS-catechol). After H2O2 was added for the second time, the GS-catechol peak disappeared and the quinone peak appeared again (Fig. 1D and Fig. S8 in Supporting information), indicating the thioether linkage is labile when treated with H2O2. These results together proved that the reaction between thiol and quinone is reversible through H2O2 treatment.

|
Download:
|
| Fig. 1. HPLC analysis of the reaction between quinone and thiol in PBS buffer (pH 7.4). (A) L-Dopa only (10 mmol/L; (B) L-Dopa was reacted with H2O2; (C) GSH (10 mmol/L) was reacted with quinone and formed GS-catechol; (D) GS-catechol was reacted with H2O2 and quinone was regenerated. | |
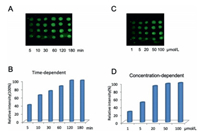
Encouraged by these results, we next explored whether this reversible reaction could be applied to microarray immobilization study. Briefly, NHS functionalized slide was prepared following previous procedure [12]. The slide was then incubated with a L-Dopa solution in NaHCO3 buffer (pH 8). After washing away the excess reagent, the L-Dopa functionalized slide was treated with H2O2 to produce quinone slides. Next, a cysteine-containing small molecule FITC-SH was used as a model compound for immobilization study. As shown in Figs. 2A and B, the fluorescence intensity gradually increased as the immobilization time increased. Around 2 h, the fluorescence intensity plateaued, indicating that the reaction was completed on the glass surface. On the other hand, concentration-dependent experiments indicated that the slide started to saturate around 20 µmol/L (Figs. 2C and D). For the negative control experiment, the quinone slide was incubated with FITC-NH2 (Fig. S14 in Supporting information). No fluorescence was observed under the same condition, indicating that no reaction occurred between amine and quinone at PBS (pH 7.4). These results also demonstrated that the reaction between qunione and thiol is highly chemoselective and efficient.
|
Download:
|
| Fig. 2. Time-dependent (A, B) and concentration-dependent (C, D) experiments of quinone slide reacted with FITC-SH. For A and B, the slides were incubated with FITC-SH (20 µmol/L) for 5, 10, 30, 60, 120 and 180 min respectively. For C and D, the slides were incubated with various concentrations of FITC-SH (1, 5, 20, 50 and 100 µmol/L) for 3 h. | |
We next investigated whether the thiol-quinone reaction could be used to immobilize molecules in a reversible manner. Two compounds, FITC-SH (Figs. S10 and S11 in Supporting information) and RhoB-SH (Figs. S12 and S13 in Supporting information), were synthesized and used for the reversible array immobilization experiment. FITC-SH was first spotted onto the quinone slide and incubated for 2 h. After washing with PBS buffer, the slide was scanned at FITC channel. As shown in Fig. 3A, bright green fluorescence could be observed, indicating successful immobilization of FITC-SH. After treatment with H2O2, the slide was washed and scanned at the same channel. No fluorescence signal was observed, indicating that the FITC-SH molecule was cleaved from the slide. Subsequently RhoB-SH was spotted on the reused slides and incubated for 2 h. After washing with PBS buffer, the slides were scanned through red channel. Bright red fluorescence could be observed, demonstrating that RhoB-SH was efficiently linked to the glass slide. After treating with H2O2, the red fluorescence disappeared, signifying RhoB-SH was removed from the slides (Figs. 3A and B). These experiments proved that the immobilization method is reversible, which could potentially provide a useful and economic tool in the microarray field.

|
Download:
|
| Fig. 3. (A) Reversible microarray immobilization of thiol-containing molecules on the quinone slides. The quinone slides were treated with FITC-SH, H2O2, RhoB-SH and H2O2 sequentially. (B) Relative fluorescent intensity of the immobilization experiment at each step. | |
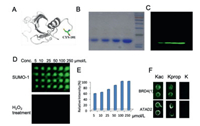
After proving that the method is suitable for immobilization of small molecules, we next investigated whether this strategy could be used for macromolecule immobilization, e.g., proteins. As a proof-of-concept experiment, we select Sumo1 protein in our model study. Sumo1 protein plays versatile roles in regulating important cellular processes such as transcription regulation and apoptosis [30]. We first mutate the C-terminal residue of Sumo1 (Val- to Cys-) to facilitate immobilization process (Fig. 4A). Subsequently we expressed, purified and fluorescently labelled the protein. After confirming mutated Sumo1 was successfully labelled by fluorescent dye (Figs. 4B and C), we spotted various concentrations of Sumo1 onto the quinone-modified slides. The protein was then incubated with the slide for 2 h. After washing with 0.5% PBST, the slide was scanned using a fluorescence scanner. As shown in Figs. 4D and E, the fluorescence intensity increased when the concentration of Sumo1 increased. The slides started to get saturated at 50 µmol/L of Sumo1. The result demonstrated that the quinone slide is capable of immobilizing macromolecules. In addition, the Sumo1 protein can be removed from the slide after treating with H2O2 (Fig. 4D). Also, we utilized Sumo1 protein without mutation as control (Fig. S15 in Supporting information), from the result, it shows that the protein without mutation cannot be immobilized on the quinone slide.
|
Download:
|
| Fig. 4. (A) Structure of Sumo-1 protein. A cysteine residue (Cys-101) was introduced on the C-terminus through site directed mutagenesis method; (B) SDS page analysis of Sumo-1 protein (5, 10, 20 and 40 µmol/L, from left to right). (C) Fluorescent gel analysis of Cy3-labeled Sumo-1 protein (5, 10, 20 and 40 µmol/L, from left to right). (D) Microarray immobilization of Cy3-labelled Sumo-1. The protein concentrations are 5, 10, 25, 50, 100 and 250 µmol/L respectively. Subsequently the slides were treated with H2O2. (E) Plot of fluorescence intensity with increasing concentrations of Sumo-1. (F) Peptides were immobilized onto microarray for bromodomain interaction study. Kac-acetylation, Kprop-propionylation, K-non-acylation. | |
Furthermore, we also synthesized three histone-derived peptides with different acylations and investigated their interactions with bromodomains. Bromodomains have emerged as attractive therapeutic targets for treating cancers [31]. As shown in Fig. 4F, the peptides with propionylation and acetylation were able to interact with bromodomains, whereas the peptide without acylation showed no binding. The experimental results also prove that the immobilization strategy can be used to fabricate peptide microarrays to investigate peptide/protein interactions.
In summary, we have successfully developed a reversible covalent microarray immobilization strategy based on the thiol-quinone reaction. The approach is highly versatile and can be used for immobilizing thiol-containing molecules, e.g., small molecules, peptides and proteins. It is noted that not every protein contains thiol residues in the sequence. However, this problem could be solved by protein engineering methods including site directed mutagenesis. Moreover, our slide can be reused and does not require the synthesis of building blocks, making it highly economical and convenient. On account of the above-mentioned prominent characteristics, it is envisioned that this reversible and versatile immobilization approach will add new and useful tools for microarray-based studies in the future.
Declaration of competing interestThe authors declare no conflict of interest.
AcknowledgmentsThe authors are grateful for the financial support from the Research Grants Council of Hong Kong (Nos. 11102719, 11304118 and 14306317), Shenzhen Basic Research Project (No. JCYJ20160601173218804), National Natural Science Foundation of China (No. 21778044, and the Shandong University of Technology Ph.D. Startup Foundation (No. 420033).
Supplementary materialsSupplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cclet.2021.06.056.
| [1] |
Z.H. Huang, L. Shi, J.W. Ma, et al., J. Am. Chem. Soc. 134 (2012) 8730-8733. DOI:10.1021/ja211725s |
| [2] |
A. Afshari, F. Nuwaysir, J. Carl Barrett, Cancer Res. 59 (2012) 4759-4763. |
| [3] |
P. Jonkheijm, D. Weinrich, M. Köhn, et al., Angew. Chem. 120 (2008) 4493-4496. DOI:10.1002/ange.200800101 |
| [4] |
Y.M. Foong, J. Fu, S.Q. Yao, M. Uttamchandani, Curr. Opin. Chem. Biol. 16 (2012) 234-242. DOI:10.1016/j.cbpa.2011.12.007 |
| [5] |
H. Sun, C.H.S. Lu, M. Uttamchandani, et al., Angew. Chem. 120 (2008) 1722-1726. DOI:10.1002/ange.200703473 |
| [6] |
S. Lata, A. Reichel, R. Brock, et al., J. Am. Chem. Soc. 127 (2005) 10205-10215. DOI:10.1021/ja050690c |
| [7] |
P.C. Lin, S.H. Ueng, M.C. Tseng, et al., Angew. Chem. 118 (2006) 4392-4396. DOI:10.1002/ange.200600756 |
| [8] |
A. Vaish, V. Silin, M.L. Walker, et al., Chem. Commun. (Camb) 49 (2013) 2685-2687. DOI:10.1039/c3cc00077j |
| [9] |
A. Girish, H. Sun, D.S.Y. Yeo, et al., Bioorg. Med. Chem. Lett. 15 (2005) 2447-2451. DOI:10.1016/j.bmcl.2005.03.079 |
| [10] |
H. Sun, S. Chattopadhaya, J. Wang, S.Q. Yao, Anal. Bioanal. Chem. 386 (2006) 416-426. DOI:10.1007/s00216-006-0511-5 |
| [11] |
G. MacBeath, S.L. Schreiber, Science 89 (2000) 1760-1763. |
| [12] |
L. Gao, S.S. Lee, J. Chen, et al., Methods Mol. Bio. 1368 (2016) 181-196. |
| [13] |
M.L. Lesaicherre, M. Uttamchandani, G.Y.J. Chen, S.Q. Yao, Bioorg. Med. Chem. Lett. 12 (2002) 2079-2083. DOI:10.1016/S0960-894X(02)00379-7 |
| [14] |
T. Pauloehrl, G. Delaittre, V. Winkler, et al., Angew. Chem. Int. Ed. 51 (2012) 1071-1074. DOI:10.1002/anie.201107095 |
| [15] |
S. Arumugam, V.V. Popik, J. Am. Chem. Soc. 133 (2011) 15730-15736. DOI:10.1021/ja205652m |
| [16] |
P.C. Lin, S.H. Ueng, M.C. Tseng, et al., Angew. Chem. Int Ed. 45 (2006) 4286-4290. DOI:10.1002/anie.200600756 |
| [17] |
B.T. Houseman, J.H. Huh, S.J. Kron, M. Mrksich, Nat. Biotechnol. 20 (2002) 270-274. DOI:10.1038/nbt0302-270 |
| [18] |
P. Wang, C.J. Zhang, G. Chen, et al., Chem. Commun. 49 (2013) 8644-8646. DOI:10.1039/c3cc43566k |
| [19] |
P. Wang, Z. Na, J. Fu, et al., Chem. Commun. (Camb) 50 (2014) 11818-11821. DOI:10.1039/C4CC03838J |
| [20] |
Wang P. L.Gao, H. Lei, et al., Methods Mol. Bio. 1518 (2017) 67-80. |
| [21] |
J. Schulte-Zweckel, F. Rosi, D. Sreenu, et al., Chem. Commun. (Camb) 50 (2014) 12761-12764. DOI:10.1039/C4CC04120H |
| [22] |
P. Kord Forooshani, B.P. Lee, J. Polym. Sci. Part A: Polym. Chem. 55 (2017) 9-33. DOI:10.1002/pola.28368 |
| [23] |
I. Putzier, P.H. Kullmann, J.P. Horn, E.S. Levitan, J. Neurophysiol. 101 (2009) 926-933. DOI:10.1152/jn.91144.2008 |
| [24] |
C. Coestentin, Chem. Rev. 108 (2008) 2145-2179. DOI:10.1021/cr068065t |
| [25] |
L. Roffe, K. Schmidt, E. Ernst, J. Clin. Oncol. 22 (2004) 4418-4424. DOI:10.1200/JCO.2004.02.034 |
| [26] |
C. Xu, K. Xu, H. Gu, et al., J. Am. Chem. Soc. 126 (2004) 9938-9939. DOI:10.1021/ja0464802 |
| [27] |
W. Ma, Y.T. Long, Chem. Soc. Rev. 43 (2014) 30-41. DOI:10.1039/C3CS60174A |
| [28] |
S. Cao, X. Peng, Curr. Org. Chem. 43 (2014) 70-76. |
| [29] |
M. Asanuma, I. Miyazaki, N. Ogawa, Neurotox Res. 5 (2003) 165-176. DOI:10.1007/BF03033137 |
| [30] |
M.P. Desterro, M.S. Rodriguez, R.T. Hay, Mol. Cell 2 (1998) 233-239. DOI:10.1016/S1097-2765(00)80133-1 |
| [31] |
L. Feng, M. Chhabra, W.H. So, Chin. Chem. Lett. 29 (2018) 1147-1150. DOI:10.1016/j.cclet.2018.05.031 |
 2022, Vol. 33
2022, Vol. 33 

