BoNT/A is the most potent toxin known to mankind and an important bio-threat agent. It is composed of a heavy chain (HC) and a light chain (LC). The HC is responsible for receptor recognition and toxin internalization, while the LC is a Zn2+ metalloprotease that selectively hydrolyzes SNAP-25 (synaptosome-associated protein-25 kDa) [1-3]. As a result of SNAP-25 cleavage, the release of acetylcholine is inhibited, leading to muscle paralysis and death.
The current treatment of BoNT/A poisoning is through antitoxin administration, but the current antitoxins are ineffective once the BoNT/A is internalized into neuromuscular cells [4], where the LC persistently cleaves SNAP-25 and results in the prolonged poisoning or even death of poison victims. Therefore, it is important to develop non-antibody drugs for the treatment of BoNT poisoning, especially drugs that are effective against the BoNT/A LC [5]. To date, some LC inhibitors, such as hydroxamate [6-8], quinolone derivatives [6, 9, 10], peptides [11-15] and other small molecules [16, 17] have been found to potently inhibit the LC of BoNT/A in vitro, but no inhibitor has been reported to improve survival rate in animal lethality assays, only few inhibitors showed modest extension of time to death of mice [18, 19]. CPI-1 [Dab(C)-RWTKCL-NH2] is a cyclic peptide linked by a disulfide bridge, has been found to be the most potent peptide inhibitor for the BoNT/A LC, but it has no detoxification activity in vivo [15]. This loss of activity is partly due to the reduction of the disulfide bridge by reducing agents in vivo.
It has been reported that cyclic peptides possess higher activity and stability compared to linear peptides [20]. The stability of easily reduced disulfide bridge-containing peptides could be improved by using disulfide bridges mimics, such as thioether (S-C)- or biscarba (C-C) bridge, triazole bridge, etc. [21-24]. In order to improve the inhibitory activity of CPI-1 in vivo against BoNT/A, we used either thioether (S-C)- or biscarba (C-C)-bridged diaminodiacid to replace the disulfide bridge of CPI-1 (Scheme 1). Based on the structure–activity of CPI-1 (unpublished data), hydrophobic and positively charged amino acids (e.g., Trp, Leu and Arg) were further added at the C-terminus of CPI-1 disulfide bond mimics (Table 1). We then determined the inhibitory activities of CPI-1 derivatives against the LC of BoNT/A, the degradation of these derivatives by trypsin and the detoxification activities of the derivatives against BoNT/A in vivo.

|
Download:
|
| Scheme 1. Synthesis route of CPI-1 disulfide bridge mimics. (a) Structures of orthogonally protected diaminodiacids. (b) Solid-phase synthesis route of CPI-1 disulfide bridge mimic by using the protected diaminodiacids. The following protecting groups for amino acid side chains were used: tert-butyl (for Thr), 2, 2, 4, 6, 7-pentamethyldihydrobenzofurane-5-sulfonyl (Pbf; for Arg) and tert-butyloxycarbonyl (Boc; for Trp, Dab and Lys). | |
|
|
Table 1 Amino acid sequence of CPI-1 and its derivatives and inhibitory activity. |
The C-S diaminodiacids X1 and X2 (Scheme 1a) were synthesized from protected L-homoserine and L-cystine as reported previously [22, 25]. The C-C-bridged diaminodiacid (X3) was prepared by using nickel-catalyzed reductive cross-coupling reaction according to the literature [23]. The allyl/allyloxycarbonyl (alloc) protecting group was employed because it remains stable during the subsequent Fmoc solid-phase peptide synthesis (SPPS) and can be easily removed by [Pd(PPh3)4]/PhSiH3. The synthesis steps, mass and nuclear magnetic resonance (NMR) spectra are shown in Figs. S1–S9 (Supporting information).
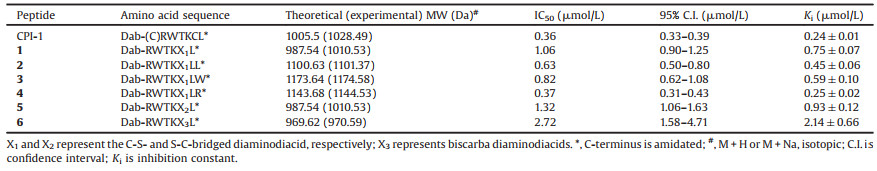
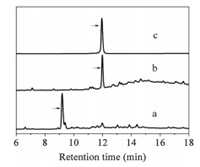
With purified diaminodiacid building blocks, we synthesized CPI-1 disulfide bridge mimics (Table 1) using solid-phase method as described previously (Scheme 1b) [22]. Fmoc-protected amino acids were assembled on Rink resin by using the coupling reagent 2-(1H-benzotriazole-1-yl)-1, 1, 3, 3-tetramethyluronium hexafluorophosphate (HBTU)/1-hydroxybenzotriazole (HOBt)/N, N-diisopropyl ethylamine (DIPEA). First, the alloc and allyl groups of diaminodiacids on the protected linear peptide were removed with [Pd(PPh3)4]/PhSiH3. The Fmoc group at the N-terminus was subsequently removed with 20% piperidine in N, N-dimethylformamide. Cyclization was then conducted by lactamization using benzotriazole-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate (PyBOP)/HOBt/N-methylmorpholine (NMM). Finally, the peptide-resin was cleaved by trifluoroacetic acid, and the crude peptide was further purified by semi-preparative high performance liquid chromatography (HPLC). The linear CPI-1 (0.5 mg/mL) was allowed to fold into the cyclic peptide CPI-1 in the NH4HCO3 buffer (0.1 mol/L, pH 8.0–8.2) at room temperature, and the cyclic peptides were then concentrated and purified by reversed-phase (C18) HPLC [26]. HPLC analyses of the cyclization products of linear CPI-1 derivative with the S-C bridge (peptide 5) are shown in Fig. 1. Other CPI-1 derivatives were also individually cyclized and the major products were purified. The SNAPtide [FITC-T(D)RIDQANQRATK(DABCYL)Nle-NH2], which is an enzymatic substrate of the BoNT/A LC used in a fluorescence resonance energy transfer (FRET)-based assay of inhibitory activity, was synthesized as described previously (Scheme S1 in Supporting information) [27]. The purity of all final cyclic peptide products were above 95%, as confirmed by HPLC with C18 column (Figs. S10–S17 in Supporting information). The mass spectra of CPI-1 and its variants are consistent with the corresponding theoretical molecular weight (Figs. S18–S25 in Supporting information). The circular dichroism (CD) spectra of CPI-1 and its variants in 0.01 mol/L phosphate buffer show random coil structure around 190 and 260 nm (Fig. S26 in Supporting information).
|
Download:
|
| Fig. 1. HPLC analysis of the products of cyclization of typical linear CPI-1 disulfide bridge mimics. (a) Linear peptide 5; (b) Cyclic products of linear peptide 5; (c) Peptide 5 purified products. Samples were applied to a Calesil ODS-100 C18 column (4.6 mm × 250 mm) and eluted with a linear gradient of 0–1 min, 5%–10% B (B is acetonitrile containing 0.1% TFA); 1–25 min, 10%–50% B; 25–28 min, 50%–95% B at a flow rate of 1 mL/min, 214 nm of wavelength. | |
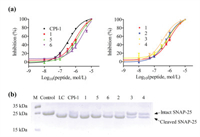
The inhibitory activities of peptides were evaluated with BoNT/A light chain 1–424 (Balc424) [27], which was expressed and purified according to the procedure described in supporting information using SNAPtide as the enzymatic substrate [28]. Dose response curves and the IC50 values of CPI-1 and its variants are shown in Fig. 2a and Table 1, respectively. After the replacement of its disulfide bridge by a C-S or C-C bridge, the IC50 values of peptides 1, 5 and 6 became 1.06, 1.32 and 2.72 μmol/L, which decreased about 1–5 fold compared to CPI-1 (IC50 = 0.36 μmol/L). On the other hand, the introduction of hydrophobic (Leu or Try) or positive charge amino acid (Arg) at the C-terminus of peptide 1 (peptides 2–4) resulted in 1–2-fold increases in the inhibitory activities. In addition, the inhibitory activities of CPI-1 and its derivatives were also assessed with full length SNAP-25 as enzymatic substrate. The peptides were pre-incubated with Balc424 at 37 ℃ for 1 h, and SNAP-25 was subsequently added and incubated for another 1 h at the same temperature. The reaction mixture was terminated by adding SDS-PAGE loading buffer and the products were analyzed by electrophoresis on a 15% polyacrylamide gel. We found that at the same concentrations, CPI-1 derivatives 3 and 4 more efficiently inhibited the BoNT/A LC-mediated cleavage of SNAP-25 compared to wild type CPI-1 and other derivatives (Fig. 2b). It should be noted that the IC50 or Ki of CPI-1 is higher than that (Ki = 39.2 nmol/L) reported in the literature [15], the reason is that the FRET substrate of CFP-SNAP-25(141–206)-YFP is more sensitive than the SANPtide [FITC–SNAP-25(189–202)-DABCYL] used in this study.
|
Download:
|
| Fig. 2. Inhibitory activities of CPI-1 disulfide bridge mimics against LC of BoNT/A. (a) Concentration-activity curves of Balc424-mediated (40 nmol/L) cleavage of SNAPtide in a FRET assay (n = 3). (b) SDS-PAGE of cleavage products of SNAP-25 (10 μmol/L) in the presence of Balc424 (1 μmol/L) and peptide inhibitors (125 μmol/L). Left to right: marker, control (SNAP-25), SNAP-25 + LC and SNAP-25 + LC + peptide. | |
To assess the stability of CPI-1 and its derivatives with disulfide bridge mimics, trypsin degradation assays were conducted. The inhibitors were allowed to incubate with trypsin (100 ng/mL) in the reaction buffer (50 mmol/L Tris-HCl, 20 mmol/L CaCl2, pH 8) at 37 ℃ for 0, 1, 2 and 3 h, respectively. HPLC analysis was used to determine the percent of degradation. As shown in Fig. 3, CPI-1 (0.25 mmol/L) was degraded by 73.4% after 8 h of incubation. The derivatives with the C-S or S-C bridge (peptides 1, 2, 3, 5) and the derivative with the C-C bridge (peptide 6) had much lower degradation, at 37.4%, 30.4%, 37.5%, 58.8% and 16.06%, respectively. Since arginine is a specific recognition site for trypsin, peptide 4 was degraded faster than other peptides. These results demonstrate that the substitution of disulfide bridge with a S-C or C-S or C-C bridge significantly improves the anti-trypsin degradation properties of the peptides.
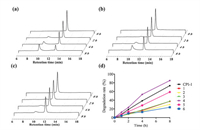
|
Download:
|
| Fig. 3. HPLC analysis of the degradation products of CPI-1 and its derivatives in the presence of trypsin. (a) CPI-1; (b) Peptide 1; (c) Peptide 6; (d) Degradation rates–time curves of CPI-1 and disulfide bridge mimics in the presence of trypsin. Analytical conditions were the same as described in Fig. 1. | |
After observing the higher anti-protease degradation resistance CPI-1 disulfide-bond mimics, we decided to further evaluate the protective activities in vivo using Kunming mice. BoNT/A [2 × median lethal dose (LD50)/mice] was mixed with CPI-1 or its derivatives (5 mg/kg), and then injected intraperitoneally into mice. The survival rates and deaths were recorded every 12 h during a time period of 72 h (Fig. 4). The results show that CPI-1 derivatives 1, 3, 4, 5 and 6 prolonged the survival time of mice. All mice in CPI-1 or the control group died 24 h after challenge, while the survival rate of peptide 1, 3, 4, 5 and 6 groups was 75%–100%. After 72 h, 25%, 25%, 25%, 25% and 37.5% of the mice receiving peptides 1, 3, 4, 5 and 6 were still alive, significantly higher than that of CPI-1 and control groups.

|
Download:
|
| Fig. 4. Survival curves of mice after the intraperitoneal injection of BoNT/A (2 × LD50) and CPI-1 or its derivatives. 2 × LD50 BoNT/A was pre-incubated with 5 mg/kg different peptides for 30 min, and was then injected intraperitoneally into mice. The death rates of mice were recorded every 12 h for 72 h. P < 0.05, **P < 0.01 vs. CPI-1 group. | |
To investigate why peptides 1, 5 and 6 have lower inhibitory activity against the BoNT/A LC, and how the introduction of hydrophobic or positive charge amino acid at the C-terminus of the peptide enhances the inhibitory activity, we compared the intermolecular interactions of Balc424 with CPI-1 and its derivatives. The crystal structure of the enzyme-CPI-1 complex shows that two salt bridge contacts exist [15], the Arg residue of CPI-1 interacts with Asp370 of Balc424, and Lys201 of CPI-1 interacts with Asp159 of Balc424. However, the salt bridge interaction between Arg of CPI-1 and Asp370 of Balc424 was not observed in Balc424-peptide 6 complex (Fig. S28A in Supporting information). CPI-1 disulfide bridge mimics have smaller size compared to wild type CPI-1, mainly due to the shorter carbon-carbon and carbon-sulfur bonds in mimics. This decrease in size may disturb salt bridge interactions and decrease the binding affinities of the peptides with the light chain of BoNT/A. For peptide 4, after the introduction of Arg at the C-terminal of peptide 1, this residue forms a salt bridge with Glu257 at Balc424 (Fig. S28B in Supporting information), which may improve the binding affinity between the two molecules.
In conclusion, we have developed a series of CPI-1 derivatives with thioether-containing and biscarba-containing diaminodiacid bridges which are more resistant to trypsin degradation. Importantly, these peptides can prolong the survival rate of mice administered with 2 × LD50 × BoNT/A. The addition of hydrophic or positive amino acid at C-terminus further improves the inhibitory activity. This research provides an efficient strategy for developing potent peptides inhibitors against BoNT/A.
Declaration of competing interestThere are no conflicts to declare.
AcknowledgmentsThis work was supported by the National Key Research and Development Project of China (2017YFC1200905). We thank Prof. Yiming Li of Hefei University of Technology, China, for helpful discussions in the synthesis experiment.
Appendix A. Supplementary dataSupplementary material related to this article canbefound, in the online version, at doi:https://doi.org/10.1016/j.cclet.2021.03.055.
| [1] |
G. Schiavo, O. Rossetto, A. Santucci, et al., J. Biol. Chem. 267 (1992) 23479-23483. DOI:10.1016/S0021-9258(18)35863-0 |
| [2] |
O. Rossetto, M. Pirazzini, C. Montecucco, Nat. Rev. Microbiol. 12 (2014) 535-549. DOI:10.1038/nrmicro3295 |
| [3] |
R.M. Benoit, D. Frey, M.M. Wieser, et al., Toxicon 107 (2015) 25-31. DOI:10.1016/j.toxicon.2015.08.002 |
| [4] |
C.N. Mayers, J.L. Holley, T. Brooks, Rev. Med. Microbiol. 12 (2001) 29-37. DOI:10.1097/00013542-200101000-00004 |
| [5] |
A.A. Adalja, E. Toner, T.V. Inglesby, N. Engl. J. Med. 372 (2015) 954-962. DOI:10.1056/NEJMra1409755 |
| [6] |
T.J. Dickerson, G.R. Smith, J.C. Pelletier, et al., Curr. Top. Med. Chem. 14 (2014) 2094-2102. DOI:10.2174/1568026614666141022095114 |
| [7] |
P. Silhar, N.R. Silvaggi, S. Pellett, et al., Bioorg. Med. Chem. 21 (2013) 1344-1348. DOI:10.1016/j.bmc.2012.12.001 |
| [8] |
R. Chauhan, V. Chauhan, P. Sonkar, et al., Mini-Rev. Med. Chem. 19 (2019) 1694-1706. DOI:10.2174/1389557519666190906120228 |
| [9] |
P.T. Bremer, M. Adler, C.H. Phung, et al., J. Med. Chem. 60 (2017) 338-348. DOI:10.1021/acs.jmedchem.6b01393 |
| [10] |
, R.C. Vieira, S.M. Ensel, et al., Bioorg. Med. Chem. Lett. 27 (2017) 675-678. DOI:10.1016/j.bmcl.2016.11.019 |
| [11] |
J.J. Schmidt, R.G. Stafford, FEBS Lett. 532 (2002) 423-426. DOI:10.1016/S0014-5793(02)03738-9 |
| [12] |
D. Kumaran, R. Rawat, M.L. Ludivico, et al., J. Biol. Chem. 283 (2008) 18883-18891. DOI:10.1074/jbc.M801240200 |
| [13] |
G. Kumar, D. Kumaran, S.A. Ahmed, et al., Acta Crystallogr. Sect. D: Struct Biol. 68 (2012) 511-520. |
| [14] |
J. Guo, J. Wang, S. Gao, et al., Sci. Rep. 5 (2015) 16981. DOI:10.1038/srep16981 |
| [15] |
D. Kumaran, M. Adler, M. Levit, et al., Bioorg. Med. Chem. 23 (2015) 7264-7273. DOI:10.1016/j.bmc.2015.10.024 |
| [16] |
H. Seki, S. Xue, M.S. Hixon, et al., Chem. Commun. 51 (2015) 6226-6229. DOI:10.1039/C5CC00677E |
| [17] |
G.E. Boldt, L.M. Eubanks, K.D. Janda, Chem. Commun. 29 (2006) 3063-3065. DOI:10.1039/b603099h |
| [18] |
L. Lin, M.E. Olson, T. Sugane, et al., J. Med. Chem. 63 (2020) 11100-11120. DOI:10.1021/acs.jmedchem.0c01006 |
| [19] |
L. Lin, M.E. Olson, L.M. Eubanks, et al., Acc. Chem. Res. 52 (2019) 2322-2331. DOI:10.1021/acs.accounts.9b00261 |
| [20] |
L.K. Buckton, M.N. Rahimi, S.R. McAlpine, Chemistry 27 (2021) 1487-1513. DOI:10.1002/chem.201905385 |
| [21] |
G.M. Fang, X.X. Chen, Q.Q. Yang, et al., Chin. Chem. Lett. 29 (2018) 1033-1042. DOI:10.1016/j.cclet.2018.02.002 |
| [22] |
H.K. Cui, Y. Guo, Y. He, et al., Angew. Chem. Int. Ed. 52 (2013) 9558-9562. DOI:10.1002/anie.201302197 |
| [23] |
T. Wang, Y.F. Kong, Y. Xu, et al., Tetrahedron Lett. 58 (2017) 3970-3973. DOI:10.1016/j.tetlet.2017.09.006 |
| [24] |
X. Li, Y. Zou, H.G. Hu, Chin. Chem. Lett. 29 (2018) 1088-1092. DOI:10.1016/j.cclet.2018.01.018 |
| [25] |
P.J. Knerr, A. Tzekou, D. Ricklin, et al., Acc. Chem. Biol. 6 (2011) 753-760. DOI:10.1021/cb2000378 |
| [26] |
F. Cai, N. Xu, Z. Liu, et al., J. Med. Chem. 61 (2018) 10198-10205. DOI:10.1021/acs.jmedchem.8b01343 |
| [27] |
M.R. Baldwin, M. Bradshaw, E.A. Johnson, et al., Protein Expr. Purif. 37 (2004) 187-195. DOI:10.1016/j.pep.2004.05.009 |
| [28] |
G.E. Boldt, J.P. Kennedy, M.S. Hixon, et al., J. Comb. Chem. 8 (2006) 513-521. DOI:10.1021/cc060010h |
 2021, Vol. 32
2021, Vol. 32