b State Key Laboratory of Bioelectronics, School of Biological and Medical Engineering, Southeast University, Nanjing 210096, China;
c Department of Cardiovascular Medicine, Zhuzhou Hospital Affiliated to Xiangya School of Medical, Central South University, Zhuzhou 412000, China;
d Hunan Shengzhou Biotechnology Ltd., Zhuzhou 412000, China;
e Guangzhou Wondfo iCubate Biotech Co., Ltd., Guangzhou 510641, China
Nucleic acid testing (NAT) is associated with high levels of specificity and sensitivity and plays an important role in the field of molecular diagnosis [1, 2], particularly with regards to the diagnosis of infectious diseases [3-9], neoplastic diseases [10, 11], cancer biomarkers [12-14], genetic mutations [15, 16], and genotyping [17], while also facilitating food safety control [18-21] and environmental monitoring [22-24]. Compared with other methods such as immune detection and microbial culture, NAT is associated with a number of significant advantages, including high levels of sensitivity and accuracy, and short operational time window; consequently, NAT can rapidly diagnose specific conditions and thus allow early therapeutic intervention [25]. The outbreak of COVID-19 in early 2020 continues to spread on a global basis. This virus has significant scope for transmission and continues to have devastating effects on a global scale. NAT has played an indispensable role in the diagnosis and control of the COVID-19 epidemic [26]. The outbreak of this epidemic is testing the medical diagnostic capability of every country; consequently, the application of NAT technologies is currently facing unprecedented demand. The huge demand for NAT during the epidemic has not been effectively met. Indeed, to speed up diagnosis, the practice of "mix-testing" has been commonly deployed. In this technique, several samples are mixed into one sample for testing, and then tested separately once abnormalities are detected.
China has repeatedly stressed that all disease control institutions at or above the county level, and general hospitals at or above the second level, should be promptly reformed so that they possess the capability to perform NAT within a short period of time. The establishment of a traditional NAT laboratory is undoubtedly a huge challenge in terms of time, cost, and manpower. Traditional NAT is demanding on the laboratory environment and operators, and unexpected errors may occur in the process [27, 28]. The emergence of point-of-care nucleic acid testing (POCNAT) provides a potential solution to this problem [29-34]. POCNAT is designed to achieve rapid NAT in areas with limited medical resources, such as emergency areas, general wards, families, fields, security check areas, and some developing countries [35]; in this regard, POCNAT uses portable devices that everyone can operate [36]. This goal can be realized by minimizing instrument size, reducing procedures, and using up-to-date information technology [37]. The COVID-19 epidemic has clearly highlighted the advantages of POCNAT, a test that is small, fast, accurate and easy [38]. In late July 2020, Premier Li Keqiang presided over an executive meeting of the State Council which pointed out that all parties should be mobilized to speed up the development and marketing of products with short detection time, high sensitivity, and simple operation. Previously, the Chinese Prime minister has twice nominated point-of-care testing (POCT) to accelerate the development of the NAT. POCNAT was clearly applicable to fill this specific niche. Previously, NAT instruments in China were mainly imported and therefore the high costs of this technology made China appear very passive in the face of major diseases. The research and development of low cost, highly sensitive, fast, and portable NAT instruments is now becoming more and more important in China and other developing countries. A few years ago, many companies had successfully developed POCT devices that could be used to detect specific pathogens with corresponding kits. Previously, there were many foreign-listed instruments, such as GeneXpert, Filmarray, Cobas Liat, and other polymerase chain reaction (PCR) amplification systems; these systems were fast and automatic but were also expensive and could not be widely used in China. The current epidemic has promoted the development and marketing of domestic POCT nucleic acid devices, such as EasyNAT, Galaxy Nano, boxarray, GenPlex, iPonatic, and AutoSAT, which have made up for the lack of POCT nucleic acid devices in China and played an important role in the epidemic.
At present, there are two issues related to POCNAT: The development of a NAT kit and the improvement of NAT instruments. The development of the NAT kit itself involves the simplification of steps [39-41], the reduction of cost [42], the improvement of sensitivity [43, 44], and reducing the processing time [45-48]. Notably, the detection kit used for POCNAT should be well matched with the detection instrument as this is conducive to the automatic integration of the instrument and the realization of rapid detection. Magnetic nanoparticles [49-52], gold nanoparticles [53-55], up-conversion nanoparticles [30], nanofibers [56], graphene oxide [57], and other new materials [58-60] and technologies [61-64], have all been used in the development of reagents. These systems can even provide us with a tool for the ultra-sensitive detection of miRNA [57, 65-67]. With regards to instrument research, the three processes required by NAT (nucleic acid extraction, target amplification, and signal detection) should be fully automated, or only require simplistic levels of manual operation. A perfect POCNAT device would have a portable size, a rapid processing time, generate accurate test results, be relatively inexpensive, involve automated operation and integrated processes, and involve multiple indicators for detection [68, 69]. Technicians should simply be able to place the sample in the instrument and then be given a simple result a short time later.
In the laboratory, NAT requires manual operation; this is very time-consuming and laborious and requires a large number of experimental carriers (e.g., tubes) and devices (e.g., pipetting guns). The application of novel carriers (chips, cartridges, and papers) will improve NAT automation. First of all, the application of such carriers significantly reduces or even avoids pollution; all of the reagents needed for the reaction are stored in the carriers, and with accurate mechanical control, the whole reaction process can be carried out automatically in the carriers, thus reducing the false-positive results caused by pollution and human intervention [70]. This not only ensures sensitivity and specificity, but also makes the NAT equipment cheaper, faster, and more portable. Moreover, the application of carriers reduces the volume of the system; which makes it more suitable for use under non-laboratory conditions. Many teams have developed disease-specific carriers to meet the needs of such experimental applications. There are also many commercial carriers under mass production, with supporting devices for NAT. Some can only detect a particular pathogen, but there are also many detection carriers that can detect many different pathogens in parallel. In some devices, nucleic acid extraction (NAE), amplification, and detection, are carried out in the same carrier, while other devices perform each of these steps in different carriers.
In this paper, we summarize the advancement of POCNAT devices based on novel experimental carriers and detection methods.
2. POCNAT methodsNAE, target amplification, and final signal detection, can be achieved stepwise by the application of a number of methods. However, the combination of different methods to create a single platform for POCT represents a particularly innovative step forward. In order to achieve a fully automatic system, a wide range of materials have been used in the extraction step of POCNAT, including silicon materials, paper-based materials, and magnetic beads [71-74]. The process of nucleic acid amplification commonly involves variable-temperature amplification [75] and isothermal amplification [76-81]; however, a number of studies have recently focused on advanced forms of digital amplification [82]. The variable form of temperature amplification involves PCR. This is a widely used technique and is one of the most powerful tools used for NAT [83]. Although conventional PCR screening is the gold standard for laboratory testing, PCR reactions require strict and complex temperature control; this increases the cost of the instrument. A PCR-based chip has recently been developed that uses independent zonal heating to reduce temperature control costs. This can involve three separate heating blocks; the liquid flows in three different temperature zones in order to achieve temperature change [84]. This modular heating method is referred to as insulated isothermal PCR (iiPCR). The PockitTM Device is a commercial NAT device that is based on iiPCR; this system has been used for the detection of a variety of RNA viruses [85-87]. The development and application of isothermal amplification technology solves the problem of temperature control in PCR [76, 88]. The isothermal amplification-based NAT device does not require an accurate temperature control system; it only requires a specific temperature. The device used for heating is simple, small in size, convenient to carry, and can hand-held. This system can amplify nucleic acids at a constant temperature with low or no power consumption, thus rendering POCT devices for nucleic acid detection less complicated, more portable, and less costly [77]. As an alternative to PCR, these systems offer significant potential in the field of POCNAT [76, 89]. At present, common isothermal amplification methods include loop-mediated isothermal amplification (LAMP) [90-93], nucleic acid sequence amplification (NASBA) [94], recombinase polymerase amplification (RPA) [95-97], helicase-dependent amplification (HDA) [98], rolling-circle amplification (RCA) [99], isothermal strand displacement amplification (iSDA) [100], and cross-priming amplification (CPA) [101]. In addition, different methods for digital nucleic acid analysis have also been developed and utilized, including digital PCR (dPCR), digital recombinase polymerase amplification (dRPA), and digital loop mediated isothermal amplification (dLAMP) [102, 103].
The signal amplified from the target sequence is converted into optical signals [104], impedance signals [105-107], chemical signals [108], electrophoretic signals [109], electrochemical signals [110-113], and magnetic signals [114], in order to provide results. There are some novel methods for signal detection. For example, the 'Spinchip', uses a spinning mechanism to achieve multi-channel detection and then applies a volumetric bar-chart for visual quantitative detection [115]. Creatively, the measurement of interfacial tension is also used to detect signals derived from digital amplification in which droplet height or shape is used as an index to measure amplification [116]. Optical detection techniques have been widely applied in POCT devices; this is because they are simple, inexpensive, convenient, and sensitive [117-119]. Optical detection usually includes colorimetric, fluorescence [120], absorbance [121, 122], chemiluminescence [123], detection, and electrochemiluminescence forms of detection. The colorimetric method is simple, direct, inexpensive and is very popular for nucleic acid testing. Colorimetric detection is based on the visible color changes produced by a specific reaction; these changes are then used for qualitative detection. Color changes are visible to the naked eye and do not require additional optical auxiliary equipment; this simplifies the detection equipment and reduces the size and cost. Colorimetric dyes are usually used as indicators. The main colorimetric dyes used at present include metal ion indicators, insertion dyes [124], and nanoparticles [125]. Signal detection can also be performed by detecting the by-products produced by a reaction [126]. However, the colorimetric method is generally only used for qualitative detection; such methods of detection are associated with low levels of sensitivity. In order to improve the accuracy of colorimetric methods and produce quantitative data, a range of optical auxiliary equipment has been considered. Charge-coupled device (CCD) cameras are often used in experiments and can be used in conjunction with convolutional neural networks of colorimetric analysis programs, such as Inception-V3 networks, one of the most widely used convolutional neural networks (CNNs) as image recognition mode; this creates a high-precision image of the color intensity of the region of interest [127].
Although highly sensitive, CCD cameras are relatively expensive. The popularity of smart phones provided researchers with a good solution. Smartphones can now be used effectively to capture the intensity of color change and calculate the gray value [128]. For example, ImageJ, a popular Java-based image processing tool, can be used to analyze the color intensity of an image [129, 130]. The color change of a product can be captured by a smartphone camera and then analyzed by ImageJ software. The drawing tool can be used to select a region of interest (ROI) on a given image and then convert the red, green, and blue (RGB) image to grayscale. In the LAMP reaction, a derivative called magnesium pyrophosphate is produced; the yield of this product increases as the amount of amplification products also increases. This derivative is a form of white precipitation; therefore, the amount of target DNA can be calculated by determining the amount of white precipitation. Such precipitation appears as turbidity in a given solution. Therefore, turbidity can be measured in real time by measuring absorbance. The photoelectric detection module of an absorbance detection system converts the received optical signal into an electric signal and then passes this signal through a series of amplification and filtering processes to obtain detection data. Fluorescence detection detects the fluorescence produced by a given reaction. There are many ways of generating fluorescence, including aptamers [131], dyes [132-134], and nanoparticles [135, 136]. Compared with colorimetric detection, fluorescence detection instruments are more complex and expensive. Fluorescence detection systems also require an excitation light source [137] and an acquisition system. The quality of the optical detection device is directly related to the quality of the entire detection process. A variety of methods have been used to improve the portability, sensitivity, accuracy, and cost, of optical detection systems [138, 139], including the use of smart phone and some homemade fluorescence detection devices. Generally, these systems are composed of three components: the excitation component, the microscopic component, and the detection component, such as a light source lens, a dichromatic mirror, and a detector, respectively. A simple fluorescence detection system would consist of a complementary metal oxide semiconductor (CMOS) camera, an excitation light source, and a reflector [140]. Portable CMOS cameras are powered by a laptop and the final results are captured and displayed on the laptop. Illumination has a significant impact on image quality. Finally, a miRNA detection system based on a light emitting diode was previously used for the detection of miR-183 template with a detection limit of approximately 4 µmol/L [141]; accurate polarization measurements were provided by the simultaneous detection of fluorescence intensity parallel to and perpendicular to the excited polarization [142].
3. ChipsMicrofluidics is characterized by the study and manipulation of fluids at the submillimeter scale and is widely used in biological detection, biological manufacturing, and tissue engineering [143-145]. The application of microfluidics in the field of biological detection can reduce the size of experiments and use a small number of samples and reagents; this technique is also less expensive and involves a shorter processing time [146]. The development and application of microfluidic technology has also enhanced the development of nucleic acid automatic detection [147, 148]. The combination of microchannels with other functional units enables the preparation, separation, detection, and other processes, to be integrated into a single chip [149]. This type of microfluidic chip, referred to as a "Lab on a Chip" (LOC), integrates all of the steps required for NAT and can even allow fully automated NAT [150]. In addition, multiple reaction units can be integrated to achieve high-throughput screening. Many researchers have tried to develop a micro-total analysis system (µTAS) based on microfluidic technology for rapid and portable NAT. Microfluidic technology has significant potential for nucleic acid quantification, mutation detection, and the construction of sequencing libraries [151]. FilmArray (BioFire, America), a commercialized instrument, uses microfluidic technology, and provides a platform for multiple POCNATs. The entire process of NAT can be integrated into a pouch chip, with all the reagents pre-stored; the entire process takes only 40-60 min. In addition, there are many types of commercial chips for NAT and many more in development. At present, chip-based NAT research can be divided into two categories, semi-integrated chips and fully integrated chips. As the name suggests, semi-integrated chips involve only one of the steps of NAT on the chip while fully integrated chips allow for the entire NAT process.
3.1. Key technologies of chip design 3.1.1. Material & fabricationIt is clearly evident that the selection of the material used to manufacture microfluidic chips, and the design of the structure, are critical decisions. For example, the selection of materials used to manufacture microfluidic chips should take into account whether the material reacts with any of the reagents, the influence of optical detection, processing difficulties, the influence of temperature, and cost. Commonly used materials are glass, quartz, monocrystalline silicon, and organic polymers. Glass and quartz exhibit good electro-osmotic and optical properties [152], although it is difficult to obtain channels with a large depth-width ratio; they are also difficult to bond. There are many different types of organic polymers; these are low cost and can pass visible light and ultraviolet light. They can be modified by chemical methods, although they are resistant to high temperatures; they also exhibit low thermal conductivity. Researchers are investigating methods that can be used to modify the surface of organic polymers. Of these polymers, polycarbonate (PC) [153, 154], poly(methyl methacrylate) (PMMA) [155], and polydimethylsilane (PDMS) [156], are the most widely used. There are many different types of new micro-processing technologies, including cast molding [157], computer numerical control (CNC) engraving machining processes [158], micro-electro-mechanical system (MEMS) technologies, and laser ablation [12]. In addition, several new sealing technologies are being developed. Chips can be designed in as many layers as needed, and often feature two [159, 160], three [46], or even five [127] layers. The bonding between each layer is important; the more layers, the more difficult the processing.
3.1.2. Driving & valvesMany steps are required to perform NAT; this increases the levels of complexity when designing a palm-sized chip. The structural design of the microfluidic chips must allow for the addition and storage of the reagents required for the reaction, as well as the isolation, flow, and automation of the various reaction steps. Microfluidic chips should also take into account the reaction requirements. It is also necessary to consider what type of driver will be used to achieve the mixing and flow of liquid within the microfluidic chips. The chips used for digital amplification also need to be specially designed to produce droplets [12, 156, 161, 162]. In order to allow for automation, fluid connection is a key factor to consider. Inside the chip, the separating, mixing, volumetric metering, aliquoting, and flow switching, of a fluid cannot be achieved without cooperation from the driving force and the valve. Researchers have designed different channels and chambers for chips according to their own individual needs by using different driving and control principles.
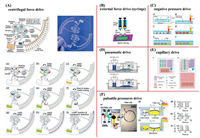
At present, the most common types of fluid drivers are centrifugal force (Fig. 1A) [163], external force (Fig. 1B) [156], negative pressure (Fig. 1C) [164], capillary force (Fig. 1E) [165], and pulsatile pressure (Fig. 1F) [166]. There are also several different types of valves, including membrane valves (Fig. 2A) [167], turning valves [168], air valves and solenoid valves (Fig. 2B) [169]. There are also some novel driving methods and valves that have become available. For example, pneumatic drives (Fig. 1D) [170] are based on electrolysis. The electrolysis process provides an inexpensive mechanism that can apply pressure to a downstream fluid using the hydrogen and oxygen generated by the electrolysis of water, thereby pumping the fluid. A centrifugal disk chip uses a laser blast valve (Fig. 2C) [127] and an ethylene-vinyl acetate copolymer (EVA) blocking valve to control the fluid. The flow rate of a chip using an active pump (such as a syringe pump [156] or pressure pump) can be adjusted independently of the chip design or fluid characteristics. However, although passive micropumps are associated with flow control problems, these pumps are still the best choice for POCT because they are simple, easy to use, and inexpensive [171].
|
Download:
|
| Fig. 1. Common drivers: (A) Centrifugal force drive. Reproduced with permission [163]. Copyright 2019, Royal Society of Chemistry; (B) external force drive (syringe). Reproduced with permission [156]. Copyright 2020, MDPI Multidisciplinary Digital Publishing Institute; (C) negative pressure drive. Copied with permission [164]. Copyright 2020, Elsevier; (D) pneumatic drive. Reproduced with permission [170]. Copyright 2013, Public Library of Science; (E) capillary drive. Copied with permission [165]. Copyright 2018, Royal Society of Chemistry; (F) pulsatile pressures drive. Reproduced with permission [166]. Copyright 2018, Royal Society of Chemistry. | |

|
Download:
|
| Fig. 2. Common valves: (A) Pressure-driven elastomeric membrane valve (PDEMV) & pressure-driven fluid guide valve (PDFGV). Reproduced with permission [167]. Copyright 2019, American Institute of Physics; (B) air valve and solenoid valve. Reproduced with permission [169]. Copyright 2019, Multidisciplinary Digital Publishing Institute; (C) laser blast valve. Copied with permission [127]. Copyright 2019, Multidisciplinary Digital Publishing Institute. | |
In some studies, reagents were loaded into chips onsite; however, the chips used for POCT and commercial chips need to be able to store reagents in advance. The pre-stored reagents in a chip are generally divided into two forms: dried reagents and liquid reagents. Dried reagents are mostly freeze-dried into powder and stored in chips at ultra-low temperature [168]. The advantage of dried reagents is that they do not require refrigeration and can be stored for long periods at room temperature. Primers are normally pre-stored in a dried form by coating the primer onto the surface of the reaction chamber at room temperature [172]. However, the cost of dry reagents increases accordingly. In some studies, primers have been deposited onto chromatographic paper and placed in the groove of a chip for amplification and fluorescence detection [173].
Usually, reagents are placed in chambers inside a chip (Fig. 3D) [174]. However, liquid reagents can also be sealed into miniaturized stick-packs (Fig. 3C) [149] or cartridges (Fig. 3B) [163] and then assembled onto a chip where it is forced into a reaction chamber to be mixed. Because of the binding of the stick-packs to the chip, the whole process of NAT can be carried out on the chip, including NAE, amplification, and detection. Sponge polyvinyl alcohol (PVA) pads can also be used to load reagents (Fig. 3A) [130]. Specifically, a stick is used to squeeze the sponge pad after the amplification reaction; the sponge is loaded with reaction reagents and samples, thus allowing the reaction solution to enter the detection area. Some chips are designed to have special chambers for storing reagents. Therefore, the size of the chip becomes larger. These large chips are also referred to as cartridges or cassettes; the reagents are stored and flow is controlled via turning valves and syringes [175].

|
Download:
|
| Fig. 3. Storage of reagents: (A) Reagents are loaded onto sponge polyvinyl alcohol pads. Copied with permission [130]. Copyright 2020, Elsevier Science & Technology; (B) reagents are sealed in cartridges [163]. Copied with permission [163]. Copyright 2019, Royal Society of Chemistry; (C) reagents are sealed in miniaturized stick-packs. Copied with permission [149]. Copyright 2020, John Wiley and Sons Inc.; (D) reagents are placed in chambers inside the chip. Reproduced with permission [174]. Copyright 2015, Royal Society of Chemistry. | |
According to specific needs and conditions, the inner surface of chips needs to be covered with a hydrophobic treatment [176]. Some novel technologies and materials are actively integrated into the fabrication of microfluidic chips. For example, three-dimensional (3D) printing technology is used for chip processing, photonic crystals are used for substrates [177], and chitosan is used for modification. The synthesis of the hydrogel used on a chip increases the sensitivity and specificity [178]. Commercial track-etched personal computer (PC) membrane is used on chips that are designed for digital amplification [82]. Chips have also been designed that feature the free-flow electrophoretic preconcentration of viral particles with thermal lysis and gel-electrophoretic NAE [179]. Another study used field effect transistors with deformed monolayer-layer graphene channels on a polystyrene substrate to detect nucleic acids [180]. Some chips that exhibit NAE functionality need to store the glass beads [181], silica beads [182], or magnetic beads [183], that are required by this process.
3.2. Semi-integrated chipsSemi-integrated chips focus on specific parts of the NAT process, including fully automated NAE. A chip features a snake-tube design for NAE [167]. All of the reagents required for this process are pre-loaded in a serpentine tube and securely isolated through a membrane valve. NAE from complex serum samples can be completed in 1 min by the application of powerful ultrasound. When air pressure is applied from the inlet, the elastic film deforms and connects the channel. The pressure-driven elastomeric membrane valve (PDEMV) in the chip uses an elastomeric membrane seal to act as a switch to control the flow of reagents. A pressure-driven fluid guide valve (PDFGV) is then used to connect the reaction chamber, reagent storage chamber, and waste water outlet, to direct the reagent to different reaction chambers according to pressure differences between the channels. The periodicity of the pulsating pressure can effectively reduce the risk of blockage in the channels. This research innovatively realizes NAE on an integrated microfluidic chip. Such chips are simple to operate, highly specific, only require short run times, and are inexpensive. However, the platform currently requires manual operation, and the design of the chip structure is too complex, thus making it difficult to shape during processing.
Compared with NAE, the chips used for nucleic acid amplification and detection can be integrated in a much easier way; consequently, there are numerous research studies on this topic. Chips based on centrifugal force control are generally disk-shaped. These disc-shaped chips do not require multiple pumps; they use a rotating motor [157] to control the centrifugal force to allow fluid to flow inside [149]. The function of the centrifugal pump is to make the fluid flow from the center to the outside diameter, while the microstructure controls the fluid flow from one chamber to another chamber [15]. The flow rate in the channel is determined by speed, the size of the connecting channel, the position of the fluidic chamber, and the viscosity of the fluid [163]. The RTISoChip (CapitalBio, China), a commercialized instrument, uses centrifugal microfluidic technology to realize multiple detection. However, this chip cannot perform NAE; it can only perform nucleic acid amplification and detection.
A study has combined "quasi" confocal laser-induced fluorescence technology, delicate microfluidic chip design, and dPCR to develop a detection device [184]. The innovative concept here is that the device uses two parallel laser beams to illuminate a 90 µm diameter droplet at the defocusing position to produce a signal with maximum fluorescence. However, the chip design and equipment manufacturing process required to make this system are relatively complex. A wearable NAT device has also been manufactured in the style of a wristband; this uses a flexible chip containing HIV-1 DNA and RPA reagents made from PDMS; this is heated by human body temperature to generate a fluorescent signal [185]. Then, a fluorescence detection system on a mobile phone was used to record the amplification results. The images were then analyzed by ImageJ to quantify the fluorescence signal intensity at different concentrations of HIV-1 DNA. Although this device has the characteristics of rapid detection, portability, and simple operation, it requires rapid nucleic acid extraction on site, and the reagent cost is high.
Chips used for digital amplification require the design of a structure that can generate droplets. A self-partitioning SlipChip has been developed for droplet generation; this system also includes dLAMP for the detection and quantification of HPV DNA [186]. The self-partitioning SlipChip is composed of two layers of glass plates. By a simple sliding step, an aqueous solution can be solidly self-segmented into individual droplets by a capillary pressure-driven flow. In addition, the design of these chips needs to move towards multiple detection capability. A silicon-based microfluidic chip with 10 parallel flow channels and a shared sample inlet was designed to perform the multiplexed detection of nucleic acids [187]. However, these chips do not have the ability to extract nucleic acids.
3.3. Fully integrated chipsFully integrated chips are bound to have more complex structural designs and valve controls than semi-integrated chips. The difficulty involved with designing and processing these chips has hindered their progress, and far less research has been carried out on fully integrated systems than semi-integrated systems.
A silicon-plastic-based disk-shaped microfluidic chip has been developed that integrates all of the components needed for sample testing on one chip [149]. This chip uses magnetic bead extraction to extract nucleic acid, and then uses real-time PCR (RT-PCR) for nucleic acid amplification detection. In addition, this chip is designed to be driven by centrifugal force and sticks-packs are used for the storage of reagents. This chip was used to detect hepatitis B virus and exhibited a limit of detection (LOD) of approximately 8 copies/reaction and a limit of quantification (LOQ) of approximately 26 copies/reaction. Although this integrated RT-PCR chip has high sensitivity for the quantitative detection of nucleic acids, its processing is complicated and costly.
Another disposable microfluidic system has also been developed; this system is based on an air valve and solenoid valve [169]. This device integrates rapid DNA extraction from clinical cervical specimens, multiple solid-phase PCR (SP-PCR), and fluorescence signal recognition, to genotype five high-risk HPV strains on a single chip. The use of air valves and solenoid valves in ordinary microfluidic chips is also very common. The pneumatic control valve undergoes deformation under the influence of air pressure and therefore controls the blockage and opening of corresponding channels. Due to the breathability of PDMS material, degassed PDMS chip blocks act in a similar way to extruded sponges in that they can suck the sample liquid into the chamber under atmospheric conditions. This chip has been used to detect 5 types of high-risk human papillomavirus (HPV) genotypes (HPV16, HPV18, HPV31, HPV33, and HPV58) and results were available in just one hour; the LOD was approximately 50 copies of HPV virus/reaction. In addition, there are some chips that use cartridges or papers for fully integrated NAT; these types of chips will be described later in this paper. Although the device integrates all the steps of nucleic acid detection on a chip, the additional pneumatic control operation is relatively complicated, which makes the operation of the detection process relatively complicated.
A two-layer structure has been created in a chip made of PDMS and bonded by a curing agent [160]. The isothermal amplification buffer, primers, probes, BST DNA polymerase, and glass powder, were placed into chambers before the two layers were bonded. The passage between the storage area and the testing area is clamped to prevent mixing. Glass powder is used to assist NAE and this process can be completed in less than 1 min. The fluorescent dye calcian is used to ensure that the detection reaction is visible to the naked eye. This microfluidic chip was able to detect L858R mutations in the gene encoding epidermal growth factor receptor (EGFRL858R) in lung adenocarcinoma NCI-H1975 cells within just 40 min. However, this chip needs to be kept at −20 ℃.
A recent invention involved the use of a chip for digital PCR based on negative pressure drive; this chip was made of PDMS material [164]. The chip was degassed in a vacuum chamber for 20 min to 0.1 kPa and a negative pressure force was used to suck reagents and oil into the microchannels to produce droplets. There is also a chip that uses a negative pressure drive, designed with a helical mixed channel and flow-focused droplet generation structure, that integrates rapid NAE and digital isothermal detection within only 60 min [188]. Although the negative pressure chip is easy to use, the processing cost is high.
4. CartridgesAlthough LOC is a promising analysis platform, it also has certain disadvantages, such as the complex fabrication steps required for microscale channels. Furthermore, these platforms are relatively expensive and require an external power supply and wire connections, thus, limiting the extent to which these systems can be reduced. The size of the chip is small, generally only the size of a human palm, but has the advantage of being thin. Due to the limitations imposed by manufacturing and processing technology, the fabrication of chips presents significant challenges. Indeed, many companies and laboratories fail to meet the relevant technical requirements. Therefore, the cartridge structure has become very common over recent years. The cartridges have the same small width and length as the chip, but its thickness is larger than that of the chip, and the volume is larger than that of the chip. In order to integrate all of the processes on the chip to achieve automation, the volume of the chip will inevitably become unavoidably larger. Cartridges can be regarded as an intermediate product of the chip. These cartridges can be regarded as large chips, although their design, manufacturing, and experimental operation, is simpler than the chip. Although cartridges are not as good as the chip in terms of size, sample number, and reagent dose, these systems are still portable, cheap, and easy to handle. In many studies, complex chips are referred to as cartridges [174]. The entire process of NAT can be integrated into a cartridge to avoid contamination caused by multiple transfers. The cartridge can also be combined with a chip or paper to realize the whole process of NAT. Optical detection on such cartridges occurs in a manner that is similar to chips; the use of smartphones is common [189]. Similar to the chip, the cartridges described in many studies also act as simple reaction vessels, but without a complex structure and a drive mechanism [190]. Pipetting is common in cartridges, where manual pipetting by laboratory personnel is replaced by mechanical arm operation; these systems often involve strict control measures to avoid errors and contamination [191].
4.1. Cartridges combined with chips or papersA small volume of fluid can be controlled by microfluidic systems in a tiny chip. A cartridge can be used for NAE and combined with on-chip detection. In a combined device, the cartridge can use a magnetic bead method for NAE, followed by a porous test chip for the simultaneous testing of multiple targets on a sample [192]. The turnaround time for a single test is approximately 30 min. Although the detection device has a short turnaround time, the chip used is too simple and associated with low levels of integration and automation.
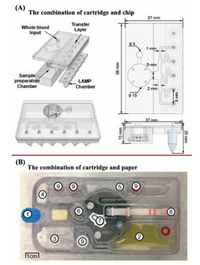
A combination of chips and cartridges also had been used to perform the entire NAT process (Fig. 4A) [193]. The microfluidic chip section has an inlet for loading blood samples and a simple microfluidic channel while the cartridge section has a sample preparation area and a LAMP area. The radius of the sample preparation chamber is 6 mm and filled with glass beads, magnetic rods, and direct buffer solution. The LAMP chamber section features six commercial standard microtubes filled with a master mixture. Syringes are then used to load samples and act as microfluidic flow pumps. However, the manufacture of the cartridge and the chip is complicated, and damage caused by insufficient craftsmanship are prone to occur. Other systems also combine a cartridge and a chip, and the feature cartridge that is used to hold waste liquid and buffer liquid [163, 181].
|
Download:
|
| Fig. 4. Cartridge combination chip or paper: (A) The combination of cartridge and chip. Copied with permission [193]. Copyright 2019, Royal Society of Chemistry; (B) the combination of cartridge and paper. Reproduced with permission [194]. Copyright 2020, Royal Society of Chemistry. | |
Target analysis with test strips (which are simple and clear), combined with the use of smart phones, can also provide quantitative results. For example, paper-based lateral flow assays can be integrated into cartridges [101]. An integrated device was previously used to detect Mycobacterium tuberculosis in sputum and consisted of a polycarbonate cartridge and a small device (Fig. 4B) [194]. This cartridge integrates pathogen lysis, NAE, DNA isothermal amplification, and a paper-based lateral flow assay. The key components of this cartridge included a four-port valve (called the Omnivalve) made from HDPE. The Omnivalve body contained 420 mg of special ceramic beads for pathogen lysis and DNA binding. Another key component is a polypropylene reaction insert which is linked to DNA amplification and a downstream lateral flow assay. Reagents are stored in a dried form in a pad and do not require refrigeration. Although the device integrates all processes, the cost, sensitivity, robustness, and other aspects, all need to be improved.
4.2. Fully integrated cartridgesFully integrated cartridges are designed with multiple chambers in which all of the reagents needed for detection are pre-stored. Under the action of an external mechanical arm, or another form of fluid driving force, all steps are carried out in the cartridge, without unnecessary manual operation, thus avoiding cross-contamination [87].
The extraction of nucleic acids using magnetic nanoparticles is a method that can be automated [195]. Some studies have integrated NAE on a cartridge and created an opportunity to perform NAE. One type of cartridge was produced by 3D printing technology; this was based on magnetic nanoparticle-based NAE and real-time fluorescence PCR for the detection of pathogens. This system also used a pipetting method to solve the upstream and downstream compatibility problems in the integrated detection process [49]. All reagents were pre-packaged into the cartridge, together with the automatic instrument controlled by computer software designed and produced by researchers. This allowed us to perform the entire process of POCNAT for pathogenic microorganisms [196]. Although the cartridge is sufficiently integrated, it requires a detection time of 2 h, and the sensitivity and portability of the device need to be improved.
The GeneXpert (Cepheid, America) is a cartridge-based nucleic acid amplification detection device that uses PCR and microfluidic technology and has a detection time of 1-2 h [197]. This commercial device has been used in many medical institutions and has been well received [198]. This system has been used for the rapid diagnosis and drug detection of Mycobacterium tuberculosis [199]. Some studies have proved that this system shows high specificity [200] but medium sensitivity for the diagnosis of extrapulmonary tuberculosis in children [201]. In addition, there are many commercial NAT devices that are based on cartridges, including Cobas Liat (Roche, America), Alere i (Abbott, America), Accula Sytem (Mesa biotech, America), Q-POC (QuantuMDX, Britain), Galaxy Nano (Igenesis, China), BoxArray (Wondfo, China), iPonatic (Sansure, China), and Tellgen (China). Most of these systems are based on microfluidic technology. However, the cost of these devices has limited their mass adoption.
5. PapersThe use of paper is also becoming a widely used approach. The recent development of paper-based microfluid technology makes it possible to achieve NAT in a simple, inexpensive, portable, fast, and effective manner in an environment with limited resources [202, 203]. Paper-based devices cannot only extract and amplify nucleic acids [204, 205], they can also perform qualitative and quantitative analysis [202, 206], and even multiple analysis [207]. These systems can be made using simple batiks or techniques. Spontaneous flow, via capillary action [208] or via the absorption of sweat on paper, does not require an external pump either. The porous structure of filter paper material can preserve DNA primers under harsh conditions, thus showing significant potential for field testing in resource-limited environments. Filter paper [209], nitrocellulose membrane [210], nylon membrane, silica membrane [211], cellulose membrane [209, 212], and Whatman filter paper, are all common paper-based materials. Cellulose paper has high mechanical strength and is particularly suitable for the imprinting of micro grooves [213]. In order to meet the specific needs of a given assay, the paper being used may need to be modified in many different ways [214]. Some studies have embedded cotton thread into paper, thus improving the detection sensitivity [215]. Chitosan is also used on nitrocellulose membranes to improve the immobilization ability of biomolecules on paper and facilitate NAE [210, 216]. By using chitosan-modified paper, the pre-treatment of samples on a vertical flow sheet eliminated the step of nuclease inactivation in the detection process; this increased the detection limit by approximately 5-fold. Because a correction fluid pen can block the pores of paper to avoid leakage and ensure the uniform flow of the sample to the reaction pad, this technology was used to draw the jet channel of the device on Whatman filter paper [129]. Similarly, hydrophobic polydimethylsiloxane prepolymers have been coated onto imprinted paper to stabilize the structure and provide a fluid barrier [213]. A polyamidoamine dendrimer was used to activate the surface of the filter paper so that it could combine with DNA quickly and specifically [209]. The Whatman FTA card is a commercial chemically treated filter paper for NAE and purification and can be installed on chips or cartridges [217]. Some studies have used self-made paper for NAE and amplification; these papers have even led to the successful amplification of DNA in untreated cells without manual intervention [218]. Functional nucleic acids, which can recognize or respond to targets of interest, have been extensively studied on paper sensors as recognition elements [219, 220]. DNA aptamers [221] are common recognition elements in POCT. Artificial synthetic xeno nucleic acids, as sensing probes, can also be used in paper POCT, and has been widely studied [220]. For example, pyrrolidine dipeptide nucleic acids, which have high levels of affinity and selectivity, can be covalently fixed to modified cellulose for the capture of target nucleic acids [110]. Colorimetric detection can be performed on paper [204, 222], as well as fluorescence detection [223]. There are also studies that combine paper with surface-enhanced Raman technology [207]. A paper-based colorimetric device has also been developed that combines with a smartphone and ImageJ software to conduct gray level analysis on images [129].
5.1. Single function paper combined with chips and cartridgesPaper is often used for NAE; for example, commercial Whatman FTA cards. A particularly creative and convenient design is an all-in-one device for the detection of multiple pathogens. This device involves commercially available Whatman FTA cards and a foldable chip [217]. This folding design integrates nucleic acid extraction, amplification, and detection, into one device, but requires manual operation and the sensitivity of visual detection will have limitations.
In addition to NAE, paper-based amplification and detection can also be integrated with chips and cartridges. For example, paper featuring dried amplification reagents and modified with chitosan, was integrated into a PMMA chip for nucleic acid amplification and detection [216]. In this example, the chip serves only as a scaffold. The sequence-specific capture of target viral RNA is one of the most commonly used methods. In this, a binding pad is modified with a viral DNA probe to capture viral RNA. This is then amplified and detected on another piece of paper that is integrated onto a chip [224]. Here, the chip serves only as a scaffold, too. Viral RNA can be successfully extracted from serum samples within 5 min while RT-LAMP can be completed within 50 min. The entire process from viral RNA extraction to detection was completed within 1 h. The disadvantage of this technique lies in the need for manual nucleic acid transfer following paper-based NAE. Moreover, for this method of amplification and fluorescence detection to be cost effective, there is a need for external equipment; furthermore, the volume is not portable. The entire process cannot be integrated and full automation is not possible.
The lateral flow assay (LFA) is one of the most widely used forms of paper-based equipment [225, 226]. The LFA uses the principle of immunochromatography, in which the test sample passes through a matrix by capillary action, thus allowing antigens to bind to the antibody to create a complex, thus enabling the detection and visualization of these complexes. A simple, portable, and economical integrated LFA device integrates paper-based nucleic acid amplification into colorimetric LFA [227]. The lateral flow strip (first layer) and the LAMP device (second layer) can then be combined to create a two-layered integrated device. The LAMP device is composed of a piece of glass fiber pad that is protected by an adhesive polyvinyl chloride (PVC) backing pad. A hand-held battery power supply system is used to amplify the target. Images in the test zones are captured by a smartphone and then processed by software on a smartphone. The common method with which to achieve multipath detection in a LFA is to arrange parallel test (T) lines on the nitrocellulose membrane; each line can then be used to identify a single biomarker. However, the finite dimension of the LFA limits its range for high-throughput detection without enlarging the LFA band. The combination of microarray and LFA technology has also led to the creation of a transverse flow microarray (LFM); this new system shows significantly improved detection throughput, although the use of colloidal gold tags limits the quantitative capability and sensitivity of the LFM. Encoded SERS nanoparticles can also be used for the detection of multiple nucleic acids on a single T line in lateral flow assay (LFA) [207]. As previously described for chips and cartridges, many designs have combined the use of transverse flow strip testing with cartridges or chips; these tests produce results directly via color changes or test lines to achieve rapid qualitative testing. However, their sensitivity and accuracy are low, and it is difficult to achieve quantification.
5.2. Microdevices for NAT based on multifunctional papersSome studies have integrated the entire NAT into papers; these papers' structures are very creative. Origami technology aims to convert the original steps of pipetting into origami; this is easier to achieve than the microfluidics by chips and the mechanical arm pipetting required by cartridges. One study used origami technology in a very innovative manner to combine sample processing, isothermal amplification, and transverse flow strip detection for the rapid, multiple detection of malaria [228]. Another test for foodborne pathogens also uses origami, but uses LAMP colorimetry (Fig. 5A) [213]; in this instance, the paper material was cellulose paper. This is because cellulose paper is very tough. The origami sections are imprinted with molds to create tiny channels and chambers. The mold is carved by a CNC milling machine. Hydrophobic polydimethylsiloxane prepolymers are then coated onto the pressed paper to stabilize the structure and provide a fluid barrier. The origami method integrates all the steps of nucleic acid detection on one piece of paper, which is easy to process. However, the manual operation of the entire process is a cumbersome and not simple enough.

|
Download:
|
| Fig. 5. Creative paper structures for whole process of NAT: (A) Origami. Copied with permission [213]. Copyright 2019, American Chemical Society; (B) sliding paper. Reproduced with permission [229]. Copyright 2019, Elsevier. | |
In addition to origami applications, there is also an innovative sliding paper application. Researchers have successfully created a plastic microdevice that contains sliding paper that fully integrates the functions of DNA extraction, LAMP, and colorimetric detection (Fig. 5B) [229]. This microdevice features three layers that allow slippage to mix samples and reagents for DNA purification, amplification, and detection in sequence. The main chamber uses an FTA card to extract and purify DNA from intact bacterial cells. Subsequently, at 65 ℃, the LAMP reagent, along with fuchsin stored in the reagent chamber, is pulled into the main chamber for DNA amplification. After 30 min, the reagent chamber where the detection reagent had stored is moved to the main chamber for result analysis. This microdevice was successfully used to screen three major foodborne pathogens in food, thus demonstrating the wide applicability of this integrated microdevice. Similarly, the device can only perform qualitative functions, not quantitative functions. The sensitivity of this device also needs to be improved.
6. Conclusion and prospectsPOCNAT devices allow the integration of multi-step NAT into specific carriers. Users only need to add samples to these devices; the remaining steps are completed automatically by the automatic control system. Results can be obtained in a very short period of time, thereby simplifying the traditional NAT. POCNAT devices liberate NAT from professional laboratories and are very applicable for field or rapid on-site testing. This has led to a revolutionary change for NAT and has broad application prospects. The core of POCT technology is to integrate NAE, amplification, and detection, into one device, and then incorporate automatic and complete detection along with results analysis. POCNAT devices that are based on enzyme amplification technology mostly use closed, disposable cartridges, chips, or papers, that can better control cross contamination. In order to facilitate the control mechanisms, many studies have carried out the extraction, amplification, and detection, of nucleic acids on different experimental carriers in turn. All processes are carried out on a single experimental carrier; this requires high levels of automation and is difficult to design and control. However, the combination of multiple carriers simplifies the manufacturing difficulty to a significant extent.
At the same time, the use of smart phones [230] and the application of certain emerging technologies, such as isothermal amplification [231, 232], nanophotonic technology [135], 3D printing technology [233, 234], microfluidic technology [235], mechanical control [236], electronic information technology [237, 238], artificial intelligence technology [239], and cloud technology [29, 187], are pushing POCT devices towards humanization and beautification [240]. Advanced digital analysis, crowd-sourced solutions, and robust user interfaces, will become an integral aspect of global health systems and personalized monitoring platforms [241]. Each technology platform has its own innovation and unique design, but also has its own advantages and disadvantages. Some focus on obtaining results quickly and in a short period of time, while others focus on multiple detection and high throughput [242]. Some allow qualitative testing while others focus more on quantitative testing. Many researchers have developed POCNAT without detection equipment; this has made created significant savings in terms of cost. However, the sensitivity of this technology has yet to be investigated. The application of smart phones can improve the sensitivity of colorimetry, at least to some extent. The sensitivity of methods involving fluorescence is much higher than those using colorimetric methods, but the cost is relatively high. Studies have tended to focus on how to improve devices to reduce costs while maintaining sensitivity. The merger of microfluidics and advanced biosensor technologies creates significant new potential for POCT, including high-throughput analysis, portability, and disposability. However, this merger also imposes technological challenges on biosensors, such as high sensitivity and selectivity requirements with sample volume orders of magnitudes smaller than those of conventional methods, false response errors due to non-specific adsorption, and the capability for integration with other necessary modules [243].
Since GeneXpert, the first test product, first went on sale in June 2010, dozens of rapid nucleic acid testing products have been launched. Although significant progress has been made with regards to POCNAT devices, it still remains a challenge to develop a portable, rapid, and simple device for automatic NAE/purification, amplification, and detection [244]. Existing commercial diagnostics focus on single-way analysis because this type of system is easy to use, easy to analyze, and suitable for mass production. However, multiplexing devices can improve the accuracy, sensitivity, and scalability, of research and diagnostic devices. The emerging optical complementary analysis and multiple contrast mechanisms enable high-reuse analysis to be implemented in field settings [241]. Moreover, there are not many instruments that can realize the complete automation of NAT. Generally, it is evident that we have only realized the partial automation of NAT [245] and still require a certain amount of manual operation, which has not achieved the real sample in-result out [246]. Therefore, there is a significant potential for us to develop POCNAT devices for personalized medicine and primary medical care. Further improvements in NAT and POCT technology [247-251] will lead to a significant expansion in their potential applications. Furthermore, it should be noted that as nucleic acid sequence analysis technology [252], genomic detection [253], liquid biopsy [254, 255], and single nucleotide polymorphism [256] research continues to mature; thus, the field of NAT will become increasingly demanding.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentsThis work was supported by the National Natural Science Foundation of China (Nos. 61901168, 61971187, 61871180, 61571187, 81902153), Zhuzhou Innovative City Construction Project (No. 2020-020), China Postdoctoral Science Foundation (No. 2018M630498), Hunan Urgency Project (No. 2020SK3005) and Education Department Outstanding Young Project of Hunan Province (No. 18B299).
| [1] |
K. Thomas, M. Gupta, S. Gaba, et al., Cureus 12 (2020) e8256. |
| [2] |
S. Singh, S. Naik, P. Sethi, et al., J. Gynecol. Surg. 36 (2020) 357-360. DOI:10.1089/gyn.2020.0045 |
| [3] |
R. Ben-Ami, A. Klochendler, M. Seidel, et al., Clin. Microbiol. Infect. 26 (2020) 1248-1253. DOI:10.1016/j.cmi.2020.06.009 |
| [4] |
J. Zhang, X. Zhang, J. Liu, et al., Int. Immunopharmacol. 88 (2020) 106861. DOI:10.1016/j.intimp.2020.106861 |
| [5] |
P.P. Nelson, B.A. Rath, P.C. Fragkou, et al., Front. Cell. Infect. Microbiol. 10 (2020) 181. DOI:10.3389/fcimb.2020.00181 |
| [6] |
J.P. Zhao, L. Chang, L.N. Wang, Eur. J. Clin. Microbiol. Infect. Dis. 38 (2019) 829-842. DOI:10.1007/s10096-019-03515-0 |
| [7] |
R.K. Yadav, D. Kumar, Y.S. Yadav, et al., J. Clin. Diagn. Res. 14 (2020) SC01-SC04. |
| [8] |
L. Tobin, L. Guerra, L. Ahouanvoeke, et al., Pan Afr. Med. J. 36 (2020) 299. |
| [9] |
A. Singhal, V.K. Sashindran, A. Aggarwal, et al., J. Clin. Diagn. Res. 14 (2020) OC01-OC04. |
| [10] |
A. Derbie, D. Mekonnen, Y. Woldeamanuel, et al., Infect. Agent. Cancer 15 (2020) 9. DOI:10.1186/s13027-020-0278-x |
| [11] |
X. Li, M. Ye, W. Zhang, et al., Biosens. Bioelectron. 126 (2019) 596-607. DOI:10.1016/j.bios.2018.11.037 |
| [12] |
B. Oliveira, B. Veigas, A.R. Fernandes, et al., Sensors 20 (2020) 1624. DOI:10.3390/s20061624 |
| [13] |
D. Sadighbayan, K. Sadighbayan, A.Y. Khosroushahi, et al., Trac-Trends Anal. Chem. 119 (2019) 115609. DOI:10.1016/j.trac.2019.07.020 |
| [14] |
Z. He, Z. Chen, M. Tan, et al., Cell Prolif. 53 (2020) e12822. |
| [15] |
G.J. Cao, J.L. Kong, Z.F. Xing, et al., Anal. Chim. Acta 1024 (2018) 123-135. DOI:10.1016/j.aca.2018.04.022 |
| [16] |
Y. Takarada, T. Kodera, K. Kobayashi, et al., J. Microbiol. Methods 177 (2020) 106062. DOI:10.1016/j.mimet.2020.106062 |
| [17] |
X.N. Liu, C. Zhang, K.W. Liu, et al., Anal. Chem. 90 (2018) 3430-3436. DOI:10.1021/acs.analchem.7b05113 |
| [18] |
J.L. Mckillip, M. Drake, J. Food Prot. 67 (2004) 823. DOI:10.4315/0362-028X-67.4.823 |
| [19] |
Y.T. Shang, J.D. Sun, Y.L. Ye, et al., Crit. Rev. Food Sci. Nutr. 60 (2020) 201-224. DOI:10.1080/10408398.2018.1518897 |
| [20] |
J. Vidic, P. Vizzini, M. Manzano, et al., Sensors 19 (2019) 1100. DOI:10.3390/s19051100 |
| [21] |
A. Poghossian, H. Geissler, M.J. Schoning, Biosens. Bioelectron. 140 (2019) 18-31. |
| [22] |
J.H. Kim, S.W. Oh, Food Control 121 (2021) 107575. DOI:10.1016/j.foodcont.2020.107575 |
| [23] |
S.F.E. Bell, L. Coffey, J. Debattista, et al., Sex Health 17 (2020) 359-367. DOI:10.1071/SH19233 |
| [24] |
R. Paul, A.C. Saville, J.C. Hansel, et al., ACS Nano 13 (2019) 6540-6549. DOI:10.1021/acsnano.9b00193 |
| [25] |
V.M. Corman, O. Landt, M. Kaiser, et al., Eurosurveillance 25 (2020) 23-30. |
| [26] |
P. Pokhrel, C.P. Hu, H.B. Mao, ACS Sens. 5 (2020) 2283-2296. DOI:10.1021/acssensors.0c01153 |
| [27] |
F. Sun, A. Ganguli, J. Nguyen, et al., Lab Chip 20 (2020) 1621-1627. DOI:10.1039/D0LC00304B |
| [28] |
T.F. Paton, I. Marr, Z. O'Keefe, et al., J. Med. Microbiol. 70 (2021) 001346. |
| [29] |
P. Wang, L.J. Kricka, Clin. Chem. 64 (2018) 1439-1452. DOI:10.1373/clinchem.2018.287052 |
| [30] |
Y. Gong, Y.M. Zheng, B.R. Jin, et al., Talanta 201 (2019) 126-133. DOI:10.1016/j.talanta.2019.03.105 |
| [31] |
Z. Li, Y. Bai, M. You, et al., Biosens. Bioelectron. 177 (2021) 112952. DOI:10.1016/j.bios.2020.112952 |
| [32] |
M. Hussain, Z. Chen, M. Lv, et al., Chin. Chem. Lett. 31 (2020) 3163-3167. DOI:10.1016/j.cclet.2020.04.038 |
| [33] |
Y. Liu, T. Li, C. Ling, et al., Chin. Chem. Lett. 30 (2019) 2211-2215. DOI:10.1016/j.cclet.2019.05.020 |
| [34] |
H. Khan, A. Khan, Y. Liu, et al., Chin. Chem. Lett. 30 (2019) 2201-2204. DOI:10.1016/j.cclet.2019.10.032 |
| [35] |
L. Zhang, B.Z. Ding, Q.H. Chen, et al., Trac-Trends Anal. Chem. 94 (2017) 106-116. DOI:10.1016/j.trac.2017.07.013 |
| [36] |
R. Peters, M. Stevenson, Clin. Microbiol. Infect. 25 (2019) 142-146. DOI:10.1016/j.cmi.2018.12.002 |
| [37] |
Q. Song, X. Sun, Z. Dai, et al., Lab Chip 21 (2021) 1634-1660. DOI:10.1039/D0LC01156H |
| [38] |
S.H. Lee, S.M. Park, B.N. Kim, et al., Biosens. Bioelectron. 141 (2019) 111448. DOI:10.1016/j.bios.2019.111448 |
| [39] |
R. Wang, R. Chen, C. Qian, et al., Sens. Actuators B: Chem. 326 (2021) 128618. DOI:10.1016/j.snb.2020.128618 |
| [40] |
S. Cai, C. Jung, S. Bhadra, et al., Anal. Chem. 90 (2018) 8290-8294. DOI:10.1021/acs.analchem.8b02062 |
| [41] |
J. Thapa, B. Maharjan, M. Malla, et al., Tuberculosis 117 (2019) 1-6. DOI:10.1016/j.tube.2019.05.004.snb.2020.128618 |
| [42] |
S. Liu, M.M. Wei, R. Liu, et al., Anal. Chim. Acta 1080 (2019) 162-169. DOI:10.1016/j.aca.2019.07.011 |
| [43] |
L.S. Yu, J. Rodriguez-Manzano, K. Malpartida-Cardenas, et al., J. Mol. Diagn. 21 (2019) 286-295. DOI:10.1016/j.jmoldx.2018.10.004 |
| [44] |
Y.F. Liu, H.P. Xu, C. Liu, et al., J. Biomed. Nanotechnol. 15 (2019) 790-798. DOI:10.1166/jbn.2019.2742 |
| [45] |
Y. Yamazaki, U. Thongchankaew-Seo, K. Nagao, et al., Lett. Appl. Microbiol. 71 (2020) 560-566. DOI:10.1111/lam.13376 |
| [46] |
G. Luo, T. Yi, Q. Wang, et al., Biosens. Bioelectron. 184 (2021) 113239. DOI:10.1016/j.bios.2021.113239 |
| [47] |
Y.X. Su, S.M. Huang, L. Hong, et al., J. Microbiol. Methods 160 (2019) 68-72. DOI:10.1016/j.mimet.2019.01.013 |
| [48] |
E. Shahbazi, H. Mollasalehi, D. Minai-Tehrani, Talanta 191 (2019) 54-58. DOI:10.1016/j.talanta.2018.08.033 |
| [49] |
H. Chen, Y.Q. Wu, Z. Chen, et al., J. Biomed. Nanotechnol. 13 (2017) 1619-1630. DOI:10.1166/jbn.2017.2478 |
| [50] |
H. Dong, C. Tang, Z. He, et al., Chin. Chem. Lett. 31 (2020) 1812-1816. DOI:10.1016/j.cclet.2020.03.002 |
| [51] |
L. Guo, T. Wang, Z. Chen, et al., Chin. Chem. Lett. 29 (2018) 1291-1295. DOI:10.1016/j.cclet.2017.11.017 |
| [52] |
L. Wang, Z.J. Liu, H.X. Cao, et al., Sens. Actuators B: Chem. 337 (2021) 129813. DOI:10.1016/j.snb.2021.129813 |
| [53] |
Y. Liu, T. Li, C. Ling, et al., Chin. Chem. Lett. 30 (2019) 2359-2362. DOI:10.1016/j.cclet.2019.10.033 |
| [54] |
Y. Lai, C. Zhang, Y. Deng, et al., Chin. Chem. Lett. 30 (2019) 160-162. DOI:10.1016/j.cclet.2018.07.011 |
| [55] |
J.Q. Huang, Y. Zhang, Z.Q. Lin, et al., Nanophotonics 8 (2019) 1495-1503. DOI:10.1515/nanoph-2019-0050 |
| [56] |
N. Wongkaew, Anal. Bioanal. Chem. 411 (2019) 4251-4264. DOI:10.1007/s00216-019-01589-5 |
| [57] |
J. Lee, Y.K. Kim, S. Lee, et al., Sens. Actuators B: Chem. 282 (2019) 861-867. DOI:10.1016/j.snb.2018.11.149 |
| [58] |
D.J. Denmark, S. Mohapatra, S.S. Mohapatra, EuroBiotech. J. 4 (2020) 184-206. DOI:10.2478/ebtj-2020-0023 |
| [59] |
J. Zhang, S. Shikha, Q.S. Mei, et al., Microchim. Acta 186 (2019) 361. DOI:10.1007/s00604-019-3449-y |
| [60] |
Y.Y. Tian, L. Zhang, L.H Wang, Biotechnol. J. 15 (2020) 1800741. DOI:10.1002/biot.201800741 |
| [61] |
S. Li, Y. Gao, Y. Ding, et al., Chin. Chem. Lett. 32 (2020) 313-318. |
| [62] |
G. Yang, Y. Lai, Z. Xiao, et al., Chin. Chem. Lett. 29 (2018) 1857-1860. DOI:10.1016/j.cclet.2018.11.030 |
| [63] |
X. Wang, F. Li, Y.R. Guo, Front. Chem. 8 (2020) 586702. DOI:10.3389/fchem.2020.586702 |
| [64] |
M. Soler, C.S. Huertas, L.M. Lechuga, Expert Rev. Mol. Diagn. 19 (2019) 71-81. DOI:10.1080/14737159.2019.1554435 |
| [65] |
M.H. Chen, R. Luo, S.H. Li, et al., Anal. Chem. 92 (2020) 13336-13342. DOI:10.1021/acs.analchem.0c02642 |
| [66] |
J. Zhang, L.L. Wang, M.F. Hou, et al., Biosens. Bioelectron. 102 (2018) 33-40. DOI:10.1016/j.bios.2017.10.050 |
| [67] |
L.P. Luo, L.L. Wang, L.P. Zeng, et al., Talanta 207 (2020) 120298. DOI:10.1016/j.talanta.2019.120298 |
| [68] |
P. Chen, C. Chen, H. Su, et al., Talanta 224 (2021) 121844. DOI:10.1016/j.talanta.2020.121844 |
| [69] |
H. Rathore, R. Biyani, H. Kato, et al., Anal. Methods 11 (2019) 4969-4976. DOI:10.1039/C9AY01476D |
| [70] |
H. Chen, Y. Wu, Z. Chen, et al., J. Biomed. Nanotechnol. 13 (2017) 1619-1630. DOI:10.1166/jbn.2017.2478 |
| [71] |
D. Lee, O. Kwon, K.H. Lee, et al., BioChip J. 13 (2019) 288-293. DOI:10.1007/s13206-019-3304-6 |
| [72] |
Z. He, C. Tang, X. Chen, et al., Mater. Express 9 (2019) 956-961. DOI:10.1166/mex.2019.1579 |
| [73] |
J.L. Chen, F. Huang, D.L. Gu, et al., Mater. Express 10 (2020) 94-101. DOI:10.1166/mex.2020.1615 |
| [74] |
Z.Q. Xiao, G.J. Yang, D. Yan, et al., Mater. Express 9 (2019) 509-516. DOI:10.1166/mex.2019.1520 |
| [75] |
G.J. Miao, L.L. Zhang, J. Zhang, et al., Anal. Chim. Acta 1108 (2020) 177-197. DOI:10.1016/j.aca.2020.01.069 |
| [76] |
C. Shi, Q. Liu, M.L. Zhou, et al., Sens. Actuators B: Chem. 222 (2016) 221-225. DOI:10.1016/j.snb.2015.08.060 |
| [77] |
L.T. Ereku, R.E. Mackay, P. Craw, et al., Anal. Biochem. 547 (2018) 84-88. DOI:10.1016/j.ab.2018.02.010 |
| [78] |
M.L. Zhang, X.D. Wang, L.Z. Han, et al., Anal. Biochem. 545 (2018) 38-42. DOI:10.1016/j.ab.2018.01.013 |
| [79] |
R. Mao, L.F. Qi, Z. Wang, et al., RSC Adv. 8 (2018) 19098-19102. DOI:10.1039/C8RA01201F |
| [80] |
C.P. Ma, Y.F. Wang, P.S. Zhang, et al., Anal. Biochem. 530 (2017) 1-4. DOI:10.1016/j.ab.2017.04.017 |
| [81] |
M. Janíková, J. Hodosy, P. Boor, et al., Microb. Biotechnol. 14 (2021) 307-316. DOI:10.1111/1751-7915.13737 |
| [82] |
X.Y. Lin, X. Huang, K. Urmann, et al., ACS Sens. 4 (2019) 242-249. DOI:10.1021/acssensors.8b01419 |
| [83] |
W. Li, A.Q. Nie, Q. Li, et al., Mater. Express 9 (2019) 484-491. DOI:10.1166/mex.2019.1511 |
| [84] |
A. Salman, H. Carney, S. Bateson, et al., Talanta 207 (2020) 120303. DOI:10.1016/j.talanta.2019.120303 |
| [85] |
J.Q. Zhang, C.R.L. Nfon, C.F. Tsai, et al., BMC Vet. Res. 15 (2019) 168. DOI:10.1186/s12917-019-1927-4 |
| [86] |
H.Y. Yin, Y.Y. Lin, C.H. Lin, et al., J. Food Saf. 39 (2019) e12690. |
| [87] |
J.J. Tsai, W.L. Liu, P.C. Lin, et al., PLoS One 14 (2019) e0218139. DOI:10.1371/journal.pone.0218139 |
| [88] |
M. Price, A. Cyrs, C.S. Sikasunge, et al., Am. J. Trop. Med. Hyg. 101 (2019) 78-83. DOI:10.4269/ajtmh.19-0121 |
| [89] |
P. Craw, W. Balachandran, Lab Chip 12 (2012) 2469-2486. DOI:10.1039/c2lc40100b |
| [90] |
A. Mohon, L.D.Y. Lee, A.G. Bayih, et al., Diagn. Microbiol. Infect. Dis. 85 (2016) 149-153. DOI:10.1016/j.diagmicrobio.2015.11.009 |
| [91] |
R. Sivakumar, V.P. Dinh, N.Y. Lee, Lab Chip 21 (2021) 700-709. DOI:10.1039/D1LC00019E |
| [92] |
J.T. Zhao, R. Feng, Virus Genes 55 (2019) 43-50. DOI:10.1007/s11262-018-1612-x |
| [93] |
Y.Z. Ling, Y.F. Zhu, H.H. Fan, et al., J. Biomed. Nanotechnol. 15 (2019) 1290-1298. DOI:10.1166/jbn.2019.2781 |
| [94] |
X. Lu, X. Shi, G. Wu, et al., Sci. Rep. 7 (2017) 1-9. DOI:10.1038/s41598-016-0028-x |
| [95] |
G. Li, F. Cong, W. Cai, et al., Aquacult. Rep. 19 (2021) 100584. |
| [96] |
A.A. El Wahed, P. Patel, M. Maier, et al., Anal. Chem. 93 (2021) 2627-2634. DOI:10.1021/acs.analchem.0c04779 |
| [97] |
J. Li, N.M. Pollak, J. Macdonald, ACS Omega 4 (2019) 11388-11396. DOI:10.1021/acsomega.9b01097 |
| [98] |
S.F. Cheung, M.F. Yee, N.K. Le, et al., Anal. Bioanal. Chem. 410 (2018) 5255-5263. DOI:10.1007/s00216-018-1178-4 |
| [99] |
B. Veigas, J. Pinto, R. Vinhas, et al., Biosens. Bioelectron. 91 (2017) 788-795. DOI:10.1016/j.bios.2017.01.052 |
| [100] |
S.N. Tammam, M.A.F. Khalil, E. Abdul Gawad, et al., Carbohydr. Polym. 164 (2017) 57-63. DOI:10.1016/j.carbpol.2017.01.051 |
| [101] |
J. Zhang, B. Di, H.B. Shan, et al., J. Food Prot. 82 (2019) 1744-1750. DOI:10.4315/0362-028X.JFP-19-156 |
| [102] |
K. Perez-Toralla, I. Pereiro, S. Garrigou, et al., Sens. Actuators B: Chem. 286 (2019) 533-539. DOI:10.1016/j.snb.2019.01.159 |
| [103] |
Y.F. Ning, X. Cui, C. Yang, et al., Anal. Chim. Acta 1055 (2019) 65-73. DOI:10.1016/j.aca.2018.12.029 |
| [104] |
T.L. Quyen, T.A. Ngo, D.D. Bang, et al., Front. Microbiol. 10 (2019) 2234. DOI:10.3389/fmicb.2019.02234 |
| [105] |
S. Sharif, Y.X. Wang, Z.Z. Ye, et al., Sens. Actuators B: Chem. 301 (2019) 127051. DOI:10.1016/j.snb.2019.127051 |
| [106] |
M. Nakano, S. Kalsi, H. Morgan, Biosens. Bioelectron. 117 (2018) 583-589. DOI:10.1016/j.bios.2018.06.063 |
| [107] |
H. Lee, J.O. Keem, H. Cho, et al., Biosens. Bioelectron. 118 (2018) 153-159. DOI:10.1016/j.bios.2018.07.050 |
| [108] |
M. Tabraue-Chavez, M.A. Luque-Gonzalez, A. Marin-Romero, et al., Sci. Rep. 9 (2019) 3696. DOI:10.1038/s41598-019-39946-0 |
| [109] |
Z.Q. Li, R.X. Ju, S. Sekine, et al., Lab Chip 19 (2019) 2663-2668. DOI:10.1039/C9LC00305C |
| [110] |
C. Srisomwat, P. Teengam, N. Chuaypen, et al., Sens. Actuators B: Chem. 316 (2020) 128077. DOI:10.1016/j.snb.2020.128077 |
| [111] |
Y.C. Liu, B.Y. Lu, Y.D. Tang, et al., Electrochem. Commun. 111 (2020) 106665. DOI:10.1016/j.elecom.2020.106665 |
| [112] |
S. Khaliliazar, L.Q. Ouyang, A. Piper, et al., ACS Omega 5 (2020) 12103-12109. DOI:10.1021/acsomega.0c00341 |
| [113] |
B. Rafique, M. Iqbal, T. Mehmood, et al., Sens. Rev. 39 (2019) 34-50. DOI:10.1108/SR-08-2017-0156 |
| [114] |
B. Tian, X.Q. Liao, P. Svedlindh, et al., ACS Sens 3 (2018) 1093-1101. DOI:10.1021/acssensors.8b00048 |
| [115] |
X.F. Wei, W. Zhou, S.T. Sanjay, et al., Anal. Chem. 90 (2018) 9888-9896. DOI:10.1021/acs.analchem.8b02055 |
| [116] |
T.H. Ulep, A.S. Day, K. Sosnowski, et al., Sci. Rep. 9 (2019) 9629. DOI:10.1038/s41598-019-46028-8 |
| [117] |
H. Zhou, J. Liu, J.J. Xu, et al., Chem. Soc. Rev. 47 (2018) 1996-2019. DOI:10.1039/C7CS00573C |
| [118] |
Y. Fan, S.F. Wang, F. Zhang, Angew. Chem. Int. Ed. 58 (2019) 13208-13219. DOI:10.1002/anie.201901964 |
| [119] |
Y. Huang, T.L. Xu, W.Q. Wang, et al., Microchim. Acta 187 (2020) 70. DOI:10.1007/s00604-019-3822-x |
| [120] |
N. Li, W.F. Zhang, Y.X. Li, et al., Trac-Trends Anal. Chem. 117 (2019) 200-214. DOI:10.1016/j.trac.2019.05.029 |
| [121] |
T.C. Li, F.J. Zhu, W. Guo, et al., RSC Adv. 7 (2017) 30446-30452. DOI:10.1039/C7RA04583B |
| [122] |
Z. Chen, T. Yang, H.W. Yang, et al., J. Biomed. Nanotechnol. 14 (2018) 198-205. DOI:10.1166/jbn.2018.2524 |
| [123] |
A. Nascetti, M. Mirasoli, E. Marchegiani, et al., Biosens. Bioelectron. 123 (2019) 195-203. DOI:10.1016/j.bios.2018.08.056 |
| [124] |
I. Banerjee, S.G. Aralaguppe, N. Lapins, et al., Lab Chip 19 (2019) 1657-1664. DOI:10.1039/C9LC00196D |
| [125] |
S. Kumar, R. Gallagher, J. Bishop, et al., Analyst 145 (2020) 6875-6886. DOI:10.1039/D0AN01098G |
| [126] |
K. Yin, V. Pandian, K. Kadimisetty, et al., Theranostics 9 (2019) 2637-2645. DOI:10.7150/thno.32224 |
| [127] |
D.Y.S. Lim, M.J. Seo, J.C. Yoo, Sensors 19 (2019) 3207. DOI:10.3390/s19143207 |
| [128] |
H.Q. Nguyen, V.D. Nguyen, H.V. Nguyen, et al., Sci. Rep. 10 (2020) 15123. DOI:10.1038/s41598-020-72095-3 |
| [129] |
V. Varsha, S. Aishwarya, S. Murchana, et al., J. Microbiol. Methods 174 (2020) 105962. DOI:10.1016/j.mimet.2020.105962 |
| [130] |
L.X. Wang, J.J. Fu, Y. Zhou, et al., Talanta 208 (2020) 120407. DOI:10.1016/j.talanta.2019.120407 |
| [131] |
M. Citartan, T.H. Tang, Talanta 199 (2019) 556-566. DOI:10.1016/j.talanta.2019.02.066 |
| [132] |
L.M. Fu, Y.N. Wang, Trac-Trends Anal. Chem. 107 (2018) 196-211. DOI:10.1016/j.trac.2018.08.018 |
| [133] |
P. Tonkrongjun, S. Phetpeng, W. Asawutmangkul, et al., Forensic Sci. Int.: Genet. 41 (2019) 168-176. DOI:10.1016/j.fsigen.2019.05.002 |
| [134] |
H. Chen, K. Liu, Z.Q. Xiao, et al., Mater. Express 10 (2020) 1463-1469. |
| [135] |
C. Tang, Z. He, H. Liu, et al., J. Nanobiotechnol. 18 (2020) 62-62. DOI:10.1186/s12951-020-00613-6 |
| [136] |
X. Pei, H. Yin, T. Lai, et al., Anal. Chem. 90 (2018) 1376-1383. DOI:10.1021/acs.analchem.7b04551 |
| [137] |
G.M. Mendes, K.G. Oliveira, J.C. Borba, et al., J. Braz. Chem. Soc. 30 (2019) 1841-1849. |
| [138] |
F. Katzmeier, L. Aufinger, A. Dupin, et al., PLoS One 14 (2019) e0220091. DOI:10.1371/journal.pone.0220091 |
| [139] |
L.A. Tortajada-Genaro, E.S. Yamanaka, A. Maquieira, Talanta 198 (2019) 424-431. DOI:10.1016/j.talanta.2019.01.124 |
| [140] |
J.Y. Jie, S.M. Hu, W.W. Liu, et al., Sensors 20 (2020) 2627. DOI:10.3390/s20092627 |
| [141] |
Z.Y. Zhang, Y.F. Wang, Y.L. Li, et al., Biotechnol. Appl. Biochem. 66 (2019) 82-90. DOI:10.1002/bab.1699 |
| [142] |
Z.J. Zhao, L. Wei, M.F. Cao, et al., Biosens. Bioelectron. 128 (2019) 91-96. DOI:10.1016/j.bios.2018.12.031 |
| [143] |
Q.F. Zhong, H.B. Ding, B.B. Gao, et al., Adv. Mater. Technol. 4 (2019) 1800663. DOI:10.1002/admt.201800663 |
| [144] |
L. Huang, E. Su, Y. Liu, et al., Chin. Chem. Lett. 32 (2020) 1555-1558. |
| [145] |
H.L. Zhu, Z. Fohlerova, J. Pekarek, et al., Biosens. Bioelectron. 153 (2020) 112041. DOI:10.1016/j.bios.2020.112041 |
| [146] |
T. Wang, Y. Deng, G. Qu, et al., Chin. Chem. Lett. 30 (2019) 1043-1050. DOI:10.1016/j.cclet.2019.01.011 |
| [147] |
Y. Yang, Y. Chen, H. Tang, et al., Small Methods 4 (2020) 1900451. DOI:10.1002/smtd.201900451 |
| [148] |
E.A. Phillips, T.J. Moehling, K.F.K. Ejendal, et al., Lab Chip 19 (2019) 3375-3386. DOI:10.1039/C9LC00506D |
| [149] |
E.L. Sciuto, S. Petralia, G. Calabrese, et al., Biotechnol. Bioeng. 117 (2020) 1554-1561. DOI:10.1002/bit.27290 |
| [150] |
J. Zhuang, J. Yin, S. Lv, et al., Biosens. Bioelectron. 163 (2020) 112291. DOI:10.1016/j.bios.2020.112291 |
| [151] |
Z.Y. Xu, Y. Qiao, J. Tu, Micromachines 10 (2019) 672. DOI:10.3390/mi10100672 |
| [152] |
W.Y. Lyu, M.C. Yu, H.J. Qu, et al., Biomicrofluidics 13 (2019) 041502. DOI:10.1063/1.5109270 |
| [153] |
Y.M. Park, S.Y. Lim, S.J. Shin, et al., Anal. Chim. Acta 1027 (2018) 57-66. DOI:10.1016/j.aca.2018.03.061 |
| [154] |
S. Su, G. Jing, M. Zhang, et al., Sens. Actuators B: Chem. 282 (2019) 60-68. DOI:10.1016/j.snb.2018.11.035 |
| [155] |
A. Naghdloo, E. Ghazimirsaeed, A. Shamloo, Sens. Actuators B: Chem. 283 (2019) 831-841. DOI:10.1016/j.snb.2018.12.084 |
| [156] |
Z.M. Zhang, S.H. Zhao, F. Hu, et al., Micromachines 11 (2020) 177. DOI:10.3390/mi11020177 |
| [157] |
B. Yang, Y.L. Fan, Y. Li, et al., Analyst 145 (2020) 3814-3821. DOI:10.1039/C9AN02572C |
| [158] |
Y.D. Ma, Y.S. Chen, G.B. Lee, Sens., Actuators B: Chem. 296 (2019) 126647. DOI:10.1016/j.snb.2019.126647 |
| [159] |
R. Li, J.W. Chen, X.J. Zhang, et al., J. Agric. Food Chem. 68 (2020) 899-906. DOI:10.1021/acs.jafc.9b06979 |
| [160] |
Y.B. Liu, Y. Zhao, Y.X. Qin, et al., RSC Adv. 6 (2016) 13399-13406. DOI:10.1039/C5RA26225A |
| [161] |
A.H. Wang, A. Abdulla, X.T. Ding, Proc. Inst. Mech. Eng. H: J. Eng. Med. 233 (2019) 683-694. DOI:10.1177/0954411919850912 |
| [162] |
J.E. Kreutz, J.S. Wang, A.M. Sheen, et al., Lab Chip 19 (2019) 1035-1040. DOI:10.1039/C8LC01223G |
| [163] |
S.J. Oh, T.S. Seo, Analyst 144 (2019) 5766-5774. DOI:10.1039/C9AN00900K |
| [164] |
H.Q. Si, G.W. Xu, F.X. Jing, et al., Sens. Actuators B: Chem. 318 (2020) 128197. DOI:10.1016/j.snb.2020.128197 |
| [165] |
A.K. Bavil, J. Kim, Analyst 143 (2018) 3335-3342. DOI:10.1039/C8AN00898A |
| [166] |
Y. Lee, D.M. Kim, Z. Li, et al., Lab Chip 18 (2018) 915-922. DOI:10.1039/C7LC01328K |
| [167] |
J.Z. Zhang, X.S. Su, J.S. Xu, et al., Biomicrofluidics 13 (2019) 034102. DOI:10.1063/1.5088552 |
| [168] |
M. Ritzi-Lehnert, R. Himmelreich, H. Attig, et al., Biomed. Microdevices 13 (2011) 819-827. DOI:10.1007/s10544-011-9552-4 |
| [169] |
C.C. Zhu, A.Z. Hu, J.S. Cui, et al., Micromachines 10 (2019) 537. DOI:10.3390/mi10080537 |
| [170] |
K. Roskos, A.I. Hickerson, H.W. Lu, et al., PLoS One 8 (2013) e69355. DOI:10.1371/journal.pone.0069355 |
| [171] |
L.F. Xu, A.Y. Wang, X.P. Li, et al., Biomicrofluidics 14 (2020) 031503. DOI:10.1063/5.0002169 |
| [172] |
H.V. Nguyen, V.D. Nguyen, H.Q. Nguyen, et al., Biosens. Bioelectron. 141 (2019) 111466. DOI:10.1016/j.bios.2019.111466 |
| [173] |
H. Wang, Z. Ma, J.X. Qin, et al., Biosens. Bioelectron. 126 (2019) 373-380. DOI:10.1016/j.bios.2018.11.011 |
| [174] |
O. Strohmeier, S. Keil, B. Kanat, et al., RSC Adv. 5 (2015) 32144-32150. DOI:10.1039/C5RA03399C |
| [175] |
L. Van Heirstraeten, P. Spang, C. Schwind, et al., Lab Chip 14 (2014) 1519-1526. DOI:10.1039/C3LC51339D |
| [176] |
Y.D. Ma, K.H. Li, Y.H. Chen, et al., Lab Chip 19 (2019) 3804-3814. DOI:10.1039/C9LC00797K |
| [177] |
K. Zhu, J.J. Chi, D.G. Zhang, et al., Analyst 144 (2019) 5413-5419. DOI:10.1039/C9AN01042D |
| [178] |
H. Lee, J. Lee, S.G. Lee, et al., Anal. Chem. 92 (2020) 5750-5755. DOI:10.1021/acs.analchem.9b05043 |
| [179] |
M. Hugle, O. Behrmann, M. Raum, et al., Analyst 145 (2020) 2554-2561. DOI:10.1039/C9AN02333J |
| [180] |
M.T. Hwang, M. Heiranian, Y. Kim, et al., Nat. Commun. 11 (2020) 1543. DOI:10.1038/s41467-020-15330-9 |
| [181] |
H. Van Nguyen, V.D. Nguyen, E.Y. Lee, et al., Biosens. Bioelectron. 136 (2019) 132-139. DOI:10.1016/j.bios.2019.04.035 |
| [182] |
R.R.G. Soares, F. Neumann, C.R.F. Caneira, et al., Biosens. Bioelectron. 128 (2019) 68-75. DOI:10.1016/j.bios.2018.12.004 |
| [183] |
R. Penchovsky, Biosens. Bioelectron. 135 (2019) 30-35. DOI:10.1016/j.bios.2019.04.014 |
| [184] |
X.R. Zhu, B.X. Liu, S.S. Su, et al., Talanta 206 (2020) 120200. DOI:10.1016/j.talanta.2019.120200 |
| [185] |
M.Q. Kong, Z.H. Li, J.G. Wu, et al., Talanta 205 (2019) 120155. DOI:10.1016/j.talanta.2019.120155 |
| [186] |
Z.Q. Yu, W.Y. Lyu, M.C. Yu, et al., Biosens. Bioelectron. 155 (2020) 112107. DOI:10.1016/j.bios.2020.112107 |
| [187] |
W.L. Chen, H.J. Yu, F. Sun, et al., Anal. Chem. 89 (2017) 11219-11226. DOI:10.1021/acs.analchem.7b02478Physics||36|18|2244|2003||| |
| [188] |
F. Hu, J. Li, Z.M. Zhang, et al., Anal. Chem. 92 (2020) 2258-2265. DOI:10.1021/acs.analchem.9b04967 |
| [189] |
H.B. Aydin, J.A. Cheema, G. Arnmanath, et al., Talanta 209 (2020) 120581. DOI:10.1016/j.talanta.2019.120581 |
| [190] |
Q. He, D.M. Yu, M.D. Bao, et al., Biosens. Bioelectron. 154 (2020) 112068. DOI:10.1016/j.bios.2020.112068 |
| [191] |
R. Tong, L.J. Zhang, C.D. Hu, et al., Micromachines 10 (2019) 204. DOI:10.3390/mi10030204 |
| [192] |
T. Yonekawa, H. Watanabe, N. Hosaka, et al., Sci. Rep. 10 (2020) 5409. DOI:10.1038/s41598-020-62109-5 |
| [193] |
H.J. Yoo, C. Baek, M.H. Lee, et al., Analyst 145 (2020) 2405-2411. DOI:10.1039/C9AN02435B |
| [194] |
H.W. Lu, R. Sakamuri, P. Kumar, et al., Lab Chip 20 (2020) 4071-4081. DOI:10.1039/D0LC00445F |
| [195] |
Z. Chen, C. Xiao, M. Tang, et al., Nanosci. Nanotechnol. Lett. 11 (2019) 1633-1643. DOI:10.1166/nnl.2019.3051 |
| [196] |
Y.L. Fang, P. Liao, S. Li, et al., Nanosci. Nanotechnol. Lett. 9 (2017) 650-659. DOI:10.1166/nnl.2017.2368 |
| [197] |
N.T.B. Phuong, N.T. Anh, N.V. Son, et al., BMC Infect. Dis. 19 (2019) 347. DOI:10.1186/s12879-019-3778-9 |
| [198] |
J. Mankotia, S. Sethi, M.A. Khan, J. Pure Appl. Microbiol. 13 (2019) 179-182. DOI:10.22207/JPAM.13.1.18 |
| [199] |
G. Durairaj, S. Rathinam, V. Vishwanathan, et al., J. Clin. Diagn. Res. 14 (2020) 19-22. |
| [200] |
T.F. Reza, T. Nalugwa, K. Farr, et al., Implement. Sci. 15 (2020) 1-11. DOI:10.1186/s13012-019-0962-7 |
| [201] |
Y.S. Seo, J.M. Kang, D.S. Kim, et al., BMC Infect. Dis. 20 (2020) 1-10. DOI:10.1186/s12879-019-4717-5 |
| [202] |
Z.D. Li, M.L. You, Y.M. Bai, et al., Small Methods 1018 (2019) 78-85. |
| [203] |
T. Akyazi, L. Basabe-Desmonts, F. Benito-Lopez, Anal. Chim. Acta 1001 (2018) 1-17. DOI:10.1016/j.aca.2017.11.010 |
| [204] |
J.J. Yang, H.B. Oh, S.H. Hwang, Sens. Actuators B: Chem. 286 (2019) 101-103. DOI:10.1016/j.snb.2019.01.122 |
| [205] |
L.Y. Liu, D.T. Yang, G.Z. Liu, Biosens. Bioelectron. 136 (2019) 60-75. DOI:10.1016/j.bios.2019.04.043 |
| [206] |
X.M. Lin, M.Y. Wu, W.B. Wang, et al., Anal. Methods 11 (2019) 1795-1801. DOI:10.1039/C8AY02715C |
| [207] |
D. Zhang, L. Huang, B. Liu, et al., Theranostics 9 (2019) 4849-4859. DOI:10.7150/thno.35824 |
| [208] |
J.W. Lee, V.D. Nguyen, T.S. Seo, BioChip J. 13 (2019) 273-279. |
| [209] |
Y.J. Song, P. Gyarmati, New Biotech. 55 (2020) 77-83. DOI:10.1016/j.nbt.2019.10.005 |
| [210] |
R.H. Tang, M. Li, L.N. Liu, et al., Cellulose 27 (2020) 3835-3846. DOI:10.1007/s10570-020-03031-x |
| [211] |
X.D. Wang, C.Y. Yan, X.K. Wang, et al., Anal. Bioanal. Chem. 412 (2020) 6927-6938. DOI:10.1007/s00216-020-02823-1 |
| [212] |
K. Ratajczak, M. Stobiecka, Carbohydr. Polym. 229 (2020) 115463. DOI:10.1016/j.carbpol.2019.115463 |
| [213] |
P.T. Trieu, N.Y. Lee, Anal. Chem. 91 (2019) 11013-11022. DOI:10.1021/acs.analchem.9b01263 |
| [214] |
R.H. Tang, L.N. Liu, S.F. Zhang, et al., Microchim. Acta 186 (2019) 521. DOI:10.1007/s00604-019-3626-z |
| [215] |
S.F. Zhang, L.N. Liu, R.H. Tang, et al., Cellulose 26 (2019) 8087-8099. DOI:10.1007/s10570-019-02677-6 |
| [216] |
E. Noviana, S. Jain, J. Hofstetter, et al., Anal. Bioanal. Chem. 412 (2020) 3051-3061. DOI:10.1007/s00216-020-02569-w |
| [217] |
K.T.L. Trinh, R.A. Stabler, N.Y. Lee, Sens. Actuators B: Chem. 314 (2020) 128057. DOI:10.1016/j.snb.2020.128057 |
| [218] |
P. Naik, S. Jaitpal, P. Shetty, et al., Sens. Actuators B: Chem. 291 (2019) 74-80. DOI:10.1016/j.snb.2019.04.044 |
| [219] |
R.D. Liu, E.M. McConnell, J.X. Li, et al., J. Mater. Chem. B 8 (2020) 3213-3230. DOI:10.1039/C9TB02584G |
| [220] |
J. Hu, K. Xiao, B.R. Jin, et al., Biotechnol. Bioeng. 116 (2019) 2764-2777. DOI:10.1002/bit.27106 |
| [221] |
Z. Guo, Y. Liu, N. He, et al., Chin. Chem. Lett. 32 (2021) 40-47. DOI:10.1016/j.cclet.2020.11.061 |
| [222] |
A. Suea-Ngam, I. Choopara, S. Li, et al., Adv. Healthc. Mater. 10 (2021) 2001755. DOI:10.1002/adhm.202001755 |
| [223] |
S.H. Lee, S.H. Lee, K. Won, et al., J. Biomed. Nanotechnol. 16 (2020) 166-178. DOI:10.1166/jbn.2020.2889 |
| [224] |
B.S. Batule, Y. Seok, M.G. Kim, Biosens. Bioelectron. 151 (2020) 111998. DOI:10.1016/j.bios.2019.111998 |
| [225] |
L. Huang, D. Zhang, L. Jiao, et al., Chin. Chem. Lett. 29 (2018) 1853-1856. DOI:10.1016/j.cclet.2018.11.028 |
| [226] |
J.H. Kim, S.W. Oh, J. Food Sci. Technol.: Mysore 56 (2019) 2576-2583. DOI:10.1007/s13197-019-03740-7 |
| [227] |
J.R. Choi, J. Hu, Y. Gong, et al., Analyst 141 (2016) 2930-2939. DOI:10.1039/C5AN02532J |
| [228] |
J. Reboud, G.L. Xu, A. Garrett, et al., Proc. Natl. Acad. Sci. U.S.A 116 (2019) 4834-4842. DOI:10.1073/pnas.1812296116 |
| [229] |
K.T.L. Trinh, T.N.D. Trinh, N.Y. Lee, Biosens. Bioelectron. 135 (2019) 120-128. DOI:10.1016/j.bios.2019.04.011 |
| [230] |
V.K. Rajendran, P. Bakthavathsalam, P.L. Bergquist, et al., Sens. Actuators B: Chem. 298 (2019) 126849. DOI:10.1016/j.snb.2019.126849 |
| [231] |
H.Q. Zhang, Y. Xu, Z. Fohlerova, et al., Trac-Trends Anal. Chem. 113 (2019) 44-53. DOI:10.1016/j.trac.2019.01.015 |
| [232] |
R. Martzy, C. Kolm, R. Krska, et al., Anal. Bioanal. Chem. 411 (2019) 1695-1702. DOI:10.1007/s00216-018-1553-1 |
| [233] |
J. Wu, Y. Liu, Y.W. Cui, et al., Biosens. Bioelectron. 142 (2019) 111508. DOI:10.1016/j.bios.2019.111508 |
| [234] |
G. Papadakis, A.K. Pantazis, M. Ntogka, et al., ACS Sens. 4 (2019) 1329-1336. DOI:10.1021/acssensors.9b00264 |
| [235] |
H.H. Shi, K.X. Nie, B. Dong, et al., Chem. Eng. J. 361 (2019) 635-650. DOI:10.1016/j.cej.2018.12.104 |
| [236] |
T. Pardy, H. Sink, A. Koel, et al., Micromachines 10 (2019) 437. DOI:10.3390/mi10070437 |
| [237] |
V.K. Rajendran, P. Bakthavathsalam, P.L. Bergquist, et al., Biosens. Bioelectron. 134 (2019) 68-75. DOI:10.1016/j.bios.2019.03.050 |
| [238] |
Y.M. Park, C.H. Kim, S.J. Lee, et al., Biosens. Bioelectron. 141 (2019) 111415. DOI:10.1016/j.bios.2019.111415 |
| [239] |
Z. Chen, C.G. Xiao, H.H. Qiu, et al., J. Biomed. Nanotechnol. 16 (2020) 1065-1081. DOI:10.1166/jbn.2020.2955 |
| [240] |
C. Wang, M. Liu, Z. Wang, et al., Nano Today 37 (2021) 101092. DOI:10.1016/j.nantod.2021.101092 |
| [241] |
A.F. Coskun, S.N. Topkaya, A.K. Yetisen, et al., Adv. Opt. Mater. 7 (2019) 1801109. DOI:10.1002/adom.201801109 |
| [242] |
S. Petralia, D. Motta, S. Conoci, Biotechnol. Bioeng. 116 (2019) 2087-2094. DOI:10.1002/bit.26987 |
| [243] |
M. Rezaei, S. Razavi Bazaz, S. Zhand, et al., Diagnostics 11 (2021) 9. |
| [244] |
M. Safavieh, C. Coarsey, N. Esiobu, et al., Crit. Rev. Biotechnol. 37 (2017) 441-458. DOI:10.3109/07388551.2016.1167667 |
| [245] |
W.N. Lee, H.J. Yoo, K.H. Nguyen, et al., Analyst 144 (2019) 6586-6594. DOI:10.1039/C9AN00896A |
| [246] |
J.H. Kang, Y.T. Kim, K. Lee, et al., Nanoscale 12 (2020) 5048-5054. DOI:10.1039/C9NR10675H |
| [247] |
H. Zhao, Q. Lin, L. Huang, et al., Nanoscale 13 (2021) 3275-3284. DOI:10.1039/D0NR08008J |
| [248] |
Y. Liu, G. Yang, T. Li, et al., Chin. Chem. Lett. 32 (2021) 1957-1962. DOI:10.1016/j.cclet.2021.01.016 |
| [249] |
S. Liu, X. He, T. Zhang, et al., Chin. Chem. Lett. (2021). DOI:10.1016/j.cclet.2021.11.051 |
| [250] |
Z. Guo, Y. Liu, N. He, et al., Chin. Chem. Lett. 32 (2021) 40-47. DOI:10.1016/j.cclet.2020.11.061 |
| [251] |
L. He, R. Huang, P. Xiao, et al., Chin. Chem. Lett. 32 (2021) 1593-1602. DOI:10.1016/j.cclet.2020.12.054 |
| [252] |
A. Lim, B. Naidenova, H. Bates, et al., J. Microbiol. Methods 159 (2019) 138-147. DOI:10.1016/j.mimet.2019.03.001 |
| [253] |
J. Ocenar, D. Arizala, G. Boluk, et al., PLoS One 14 (2019) e0218868. DOI:10.1371/journal.pone.0218868 |
| [254] |
M. Tabata, Y. Miyahara, J. Photopolym. Sci. Technol. 32 (2019) 97-100. DOI:10.2494/photopolymer.32.97 |
| [255] |
M. Tabata, Y. Miyahara, J. Mater. Chem. B 7 (2019) 6655-6669. DOI:10.1039/C9TB00718K |
| [256] |
M. Varona, J.L. Anderson, Anal. Chem. 91 (2019) 6991-6995. DOI:10.1021/acs.analchem.9b01762 |
 2021, Vol. 32
2021, Vol. 32 
