Carbon nanodots (CNDs), which have a typical size of < 10 nm, are a class of zero-dimension photoluminescent carbonaceous nanomaterials [1, 2]. CNDs have attracted tremendous attention due to their excellent features that are favorable for biomedical applications, such as tunable photoluminescence, anti-photobleaching ability, excellent water dispersibility endowed by the hydrophilic groups on the surface, modifiability contributed by the functional groups such as the amine or carboxyl group, facial preparation process (e.g., hydrothermal/solvothermal method, microwave-assisted approach, and electrochemical method), abundant source materials (including small molecules, polymers, microorganisms, and bulk carbon), and low cost [3-25]. Owing to the above-mentioned attractive properties, CNDs have been widely used in biosensing, bioimaging, and drug delivery [26-45]. However, the applications of most CNDs are sometimes hindered by their excitation-dependent emission property [6, 7, 11, 29, 46-50] that can cause fluorescence overlap with other fluorescent probes.
On the other hand, lysosome is a unique organelle that plays an important role in various cellular processes, including plasma membrane repair, cell apoptosis, autophagy, energy metabolism, cell signaling, and cell death [51-53]. Specific imaging of lysosomes is essential for investigating the above cellular behaviors. Among various techniques, the photoluminescence imaging technique is a widely used method due to the merits of easy operation, high sensitivity, and fast response [54-56]. Up to date, several strategies have been developed for the photoluminescence imaging of lysosomes, such as the combination of photoluminescent nanomaterials and lysosome-targeting molecules (e.g., morpholine-modified CNDs and ruthenium complex-modified CNDs) [57-59] and the amine modification/termination of photoluminescent nanomaterials [60-68]. Particularly, the feasibility of the amine-based approach for lysosomal targeting has been extensively proved recently. The lysosome-targeting mechanism of the approach can be attributed to the acidotropic effect of the weakly basic amine group [62, 63, 65], which can endow the amine-modified materials with the ability to accumulate in the acidic lysosome (pH 4.5-5.0). However, the current lysosome-targetable CNDs with the amine surface are confronted with the limitations of the big size (such as the lysosome-targeting CNDs with the size of 50-90 nm synthesized by Tong et al. [68]) that may result in low accuracy and resolution of fluorescence imaging, complicated preparation process because of the extra surface modification [60], low photoluminescence quantum yield (PLQY) of ≤ 20% [61-65], and multicolor emission [62, 66] that will induce emission overlap with other dyes. Therefore, it is highly desirable to develop a simple yet efficient strategy for the preparation of lysosome-targetable CNDs with favorable size, high PLQY, and single-color fluorescence.
In this work, ultrasmall CNDs (~2.2 nm) with very bright green fluorescence emission were prepared via a one-step hydrothermal method. The synthesis is easy, straightforward, and can be easily reproduced. More importantly, the CNDs have an ultrahigh PLQY of 80% and inherent amine-terminated surface, and can realize excellent lysosomal imaging without additional modification of a lysosome-targeting ligand. In addition, the ultrasmall size of the CNDs can endow them with high imaging accuracy and resolution. Therefore, this research overcomes some important shortcomings of the current lysosome-targetable CNDs. Additionally, we explain in this paper the photoluminescence mechanism of the CNDs with single color emission. We also demonstrate the lysosomal imaging performance and biocompatibility of the as-synthesized CNDs.
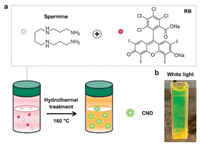
The amine-terminated green-emitting CNDs with a high PLQY of ~80% were prepared by a simple hydrothermal reaction at 160 ℃ between the amine-containing spermine and rose bengal (RB) molecules for 12 h (Fig. 1a). The as-synthesized CND aqueous suspension was yellowish-green in appearance (Fig. 1b). TEM image presents the spherical shape of the CNDs (Fig. 2a) with an ultrasmall average size of ~2.2 nm (Fig. 2b). The detected hydrodynamic diameter of the CNDs was around 12.8 nm (Fig. S1 in Supporting information). Meanwhile, the CNDs exhibited excellent aqueous stability, which was proved by the similar hydrodynamic sizes within 7 days (Fig. S2 in Supporting information). Then, the structure and composition of the CNDs were analyzed by electron diffraction and X-ray photoelectron spectroscopy (XPS). The selected area electron diffraction (SAED) pattern in Fig. S3 (Supporting information) implied the amorphous nature of the CNDs. Only three elemental peaks including C 1s, N 1s, and O 1s could be detected in the XPS spectrum of the CNDs (Fig. 2c). The corresponding high-resolution peaks in Figs. 2d-f showed the presence of various chemical bonds including the C—N—C and C—N—H bonds (from the spermine molecule), and the C—O, C=O and C=C bonds (from the RB molecule), suggesting the successful conjugation between spermine and RB during the hydrothermal process. Meanwhile, the peak at 1580 cm-1 in the Fourier transform infrared (FTIR) spectrum of CNDs (marked by the red arrow in Fig. 2g) was attributed to the vibration of the amide bond, further suggesting that the conjugation of spermine with RB was through the reaction between the amine group of spermine and the carboxyl group of RB. Besides, the absorption ranging from 3300 cm-1 to 3500 cm-1 in the FTIR spectrum (Fig. 2g) and the peak at 400.5 eV in the XPS spectrum (Fig. 2e) of CNDs were contributed by the unreacted amine groups of CNDs, which endows the CNDs with surface modifiability for further applications. Additionally, the as-prepared CNDs were characterized by 1H nuclear magnetic resonance (1H NMR) spectroscopy. As revealed in Fig. 2h, the spermine molecule possessed the characteristic chemical shifts of the amine group ranging from 1.6 ppm to 1.7 ppm. For the CNDs, the characteristic signals of the amine group were decreased, implying the reaction between the spermine and RB. The residual signals at 1.6-1.7 ppm in the 1H NMR spectrum of CNDs (Fig. 2h) could be explained by the unreacted amine groups, further confirming the FTIR results.
|
Download:
|
| Fig. 1. (a) Schematic illustration of the molecular structures of spermine and RB and the preparation process of the green-emitting CNDs. (b) Photograph of the as-prepared CNDs dispersed in water under white light exposure. | |

|
Download:
|
| Fig. 2. TEM image (a), corresponding size distribution (b), and XPS spectrum (c) of CNDs. High-resolution XPS peaks of C 1s (d), N 1s (e) and O 1s (f) of CDs. Fourier-transform infrared (FTIR) spectra (g) and 1H NMR spectra (h) of spermine, RB and CNDs. | |
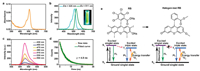
Next, the photoluminescence property of the CNDs was evaluated. The as-prepared CNDs showed a significant absorption peak at 511 nm (Fig. 3a) and exhibited strong green photoluminescence under 365 nm light exposure (Fig. 3b). The maximal photoluminescence excitation and emission peaks of the CNDs were located at 511 and 525 nm, respectively (Fig. 3b). Adopting the quinine sulfate with a PLQY of 54% as a reference, the relative PLQY of the CNDs was calculated to be as high as ~80%, which was similar to the absolute PLQY value of 76.4% detected by an Edinburgh FLSP920 fluorescence spectrophotometer. Meanwhile, the PLQY of the CNDs obtained via the 12 h of hydrothermal reaction was higher than those of the products collected via the hydrothermal reaction of 4 h (21.5%), 6 h (57.9%), and 24 h (71.6%) (Table S1 in Supporting information), indicating that 12 h might be the optimal reaction time. Compared to the multicolor fluorescence emission of most CNDs, the CNDs synthesized in this work showed the property of excitation-independent emission (Fig. 3c), which can avoid the inconvenience caused by the fluorescence overlap when the CNDs are to be used together with other fluorescent reagents/materials for fluorescence imaging. The photoluminescence lifetime of the green-emitting CNDs was measured to be 4.9 ns (Fig. 3d), which was longer than that of the RB molecule (0.6 ns) [53]. The photoluminescence mechanism of the CNDs may be explained by the changed molecular structure of RB due to the high-temperature-induced halogen atom loss during the hydrothermal reaction (Fig. 3e). For the RB molecule, it contains eight halogen atoms (4Cl and 4I), which dramatically decreases its photoluminescence efficiency (PLQY of RB: ~1%) by promoting the formation of the triplet state. After the hydrothermal reaction, the RB molecule lost all of its halogen atoms (Fig. 2c) and was changed into a structure like that of the fluorescein molecule (Fig. S4 in Supporting information) which also emits green fluorescence (Fig. S5 in Supporting information). The decrease of the heavy atoms (i.e., I) can lead to the reduced possibility to form the triplet state, thus contributing to the enhanced fluorescence efficiency and lifetime [69, 70]. The above favorable photoluminescence properties of the CNDs are important for their successful application in fluorescence imaging.
|
Download:
|
| Fig. 3. (a) Ultraviolet-visible (UV-vis) spectrum of the CND aqueous suspension. (b) Fluorescence excitation and emission spectra of the CND aqueous suspension. Inset: Photograph of the CND aqueous suspension under the 365 nm light illumination. (c) Fluorescence emission spectra of the CND aqueous suspension collected at various excitation wavelengths. (d) Time-resolved fluorescence decay result and fitted curve of the CND aqueous suspension. (e) Schematic illustration of the molecular structures and energy level diagrams of RB and halogen-lost RB moiety. "FL" indicates "fluorescence", and "PL" indicates "photoluminescence". | |
Taking advantage of their highly strong single-color fluorescence emission, the CNDs were utilized for cell imaging. MCF-7 (a human breast cancer cell line) cells were chosen as a representative cell model. As displayed in Fig. 4a, the CNDs could be sufficiently internalized by the cells after an incubation period of 90 min. The green dots from the CNDs well overlap with the red dots from the commercial lysosomal dye Lyso-Tracker Red (abbreviated as LT-Red in the figure), because the yellow signals represent the overwhelming majority in the overlay channel. The calculated Pearson correlation coefficient (PCC) is 0.81, exhibiting the ability of the CNDs for specific lysosome imaging. The PCC value was not very high, which was possibly because of the different optimal focal planes of the two reagents in confocal microscopic imaging. Furthermore, the CNDs were still distributed in the lysosomes when the staining time was increased to 48 h, displaying their excellent lysosome-targeting performance, including the high staining stability and negligible impact on lysosomal morphology, which ensure the satisfactory application of the CNDs in long-term lysosomal tracking. The working mechanism of the CNDs for lysosomal imaging was tentatively explained as follows (and was also depicted in Fig. 4b): First, the ultrasmall CNDs can be endocytosed into the cell, followed by the formation of the endosome. Then, the endosome transports the CNDs to the lysosome. Finally, the CNDs with the weakly basic amine groups (provided by the spermine molecule) accumulate in the lysosome due to the acidic lysosomal lumen (pH 4.5-5.5), realizing the specific imaging of the lysosome. Further, the lysosomal imaging ability of the CNDs was also investigated using two other cell lines including the A549 (a human lung cancer cell line) cells and HeLa (an adenocarcinoma cervical cancer cell line) cells. As shown in Figs. S6 and S7 (Supporting information), excellent colocalization of the green-emitting CNDs and the red-emitting LT-Red was observed in both A549 and HeLa cells, indicating the universal lysosomal staining capability of the CNDs.

|
Download:
|
| Fig. 4. (a) Confocal fluorescence images of the MCF-7 cells, which were treated with the as-prepared CNDs (50 μg/mL) for different time periods. Before imaging, the CND-treated cells were stained with the Lyso-Tracker Red (LT-Red) dye for the colocalization analysis. (b) Scheme illustrating the working mechanism of the CNDs in lysosomal imaging. Relative viabilities of the MCF-7 cells incubated with different concentrations of the CNDs for 24 h (c) or 48 h (d). | |
To better demonstrate the application potential of the CNDs as a lysosomal imaging probe, the long-term cytotoxicity of the CNDs was investigated. As presented in Fig. 4c, negligible cell death was detected after the culture of the MCF-7 cells with the CNDs at a high concentration of 800 μg/mL for 24 h. Besides, the CND (800 μg/mL)-treated MCF-7 cells had a relative viability of >80% even when the treatment time was prolonged to 48 h (Fig. 4d). We also tested the cytotoxicity of the as-prepared CNDs in the HEK293 (a human kidney cell line) cells. As showed in Fig. S8 (Supporting information), the HEK293 cells displayed >80% viability after the treatment of 800 μg/mL CNDs for 48 h, further ensuring the biosafety of the CNDs. The above results indicated the low cytotoxicity of the CNDs in different cell lines (including the cancer cell line and the normal cell line), which is highly beneficial for the safe use of the CNDs for lysosome-related studies.
In summary, this work reports the successful synthesis of single-color photoluminescent CNDs through a hydrothermal reaction between spermine and RB. The CNDs had the features of amine termination, excellent water dispersity, ultrasmall size (~2.2 nm), green photoluminescence, excitation-independent emission, and a high PLQY of ~80%. The photoluminescence mechanism of the CNDs was also investigated, and it was pointed out that the halogen atom loss during the hydrothermal reaction was the main cause for the very high PLQY of the obtained CNDs. Moreover, compared to other fluorescent nanoparticles, the as-prepared CNDs possess many merits. For example, the CNDs have the advantages of low cytotoxicity and low economic cost in comparison with the semiconductor quantum dots. Compared to the upconversion nanoparticles and noble metal nanoclusters which usually have low PLQYs, the high PLQY endows the CNDs with the convenience in fluorescence imaging. Meanwhile, the CNDs display the advantages of environmentally benign, simple preparation, excellent aqueous stability, and facile surface functionalization when they are in comparison to the polymer nanodots. Owing to the presence of the weakly basic surface amine groups, the CNDs could be efficiently internalized into cells and specifically accumulate in the lysosomes for realizing high-quality lysosomal imaging. We have also demonstrated that the CNDs possessed the merits of long-term lysosomal staining stability, and, negligible disturbance to the lysosomal morphology. It is believed that the CNDs may hold great promise for lysosome-related cell biology studies, and the halogen atom loss-based CND preparation strategy will also promote the development of novel CNDs for desired applications.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentThis work was supported by the National Natural Science Foundation of China (No. 21673037).
Appendix A. Supplementary dataSupplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.cclet.2021.03.061.
| [1] |
F. Arcudi, L. Dordevic, M. Prato, Acc. Chem. Res. 52 (2019) 2070-2079. DOI:10.1021/acs.accounts.9b00249 |
| [2] |
X.D. Zhang, X.K. Chen, F.G. Wu, Carbon nanodots for cell imaging, in: F.G. Wu (Ed. ), Fluorescent Materials for Cell Imaging, Springer, Singapore, Singapore, 2020, pp. 49-75.
|
| [3] |
Y.Q. Sun, H.Y. Qin, X. Geng, et al., ACS Appl. Mater. Interfaces 12 (2020) 31738-31744. DOI:10.1021/acsami.0c05005 |
| [4] |
X.W. Hua, Y.W. Bao, F.G. Wu, ACS Appl. Mater. Interfaces 10 (2018) 10664-10677. DOI:10.1021/acsami.7b19549 |
| [5] |
X.W. Hua, Y.W. Bao, J. Zeng, F.G. Wu, ACS Appl. Mater. Interfaces 11 (2019) 32647-32658. DOI:10.1021/acsami.9b09590 |
| [6] |
J.J. Yang, X.D. Zhang, Y.H. Ma, et al., ACS Appl. Mater. Interfaces 8 (2016) 32170-32181. DOI:10.1021/acsami.6b10398 |
| [7] |
S. Chen, T.T. Sun, M. Zheng, Z.G. Xie, Adv. Funct. Mater. 30 (2020) 2004680. DOI:10.1002/adfm.202004680 |
| [8] |
M.H. Lan, L. Guo, S.J. Zhao, et al., Adv. Therap. 1 (2018) 1800077. DOI:10.1002/adtp.201800077 |
| [9] |
H.F. Liu, J. Yang, Z.H. Li, et al., Anal. Chem. 91 (2019) 9259-9265. DOI:10.1021/acs.analchem.9b02147 |
| [10] |
S. Sun, Q. Chen, Z.D. Tang, et al., Angew. Chem. Int. Ed. 59 (2020) 21041-21048. DOI:10.1002/anie.202007786 |
| [11] |
G. Gao, Y.W. Jiang, H.R. Jia, J.J. Yang, F.G. Wu, Carbon 134 (2018) 232-243. DOI:10.1016/j.carbon.2018.02.063 |
| [12] |
J.J. Yang, G. Gao, X.D. Zhang, et al., Carbon 146 (2019) 827-839. DOI:10.1016/j.carbon.2019.02.040 |
| [13] |
X. Zhang, J.M. Wang, J. Liu, et al., Carbon 115 (2017) 134-146. DOI:10.1016/j.carbon.2017.01.005 |
| [14] |
C.X. Wang, Z.Z. Xu, H. Cheng, et al., Carbon 82 (2015) 87-95. DOI:10.1016/j.carbon.2014.10.035 |
| [15] |
N. Wang, Y.T. Wang, T.T. Guo, et al., Biosens. Bioelectron. 85 (2016) 68-75. DOI:10.1016/j.bios.2016.04.089 |
| [16] |
S.H. Li, D. Amat, Z.L. Peng, et al., Nanoscale 8 (2016) 16662-16669. DOI:10.1039/C6NR05055G |
| [17] |
T. Feng, X.Z. Ai, G.H. An, P.P. Yang, Y.L. Zhao, ACS Nano 10 (2016) 4410-44120. DOI:10.1021/acsnano.6b00043 |
| [18] |
Y.B. Liu, L. Zhou, Y.N. Li, R.P. Deng, H.J. Zhang, Nanoscale 9 (2017) 491-496. DOI:10.1039/C6NR07123F |
| [19] |
M.H. Sun, C. Liang, Z. Tian, et al., J. Phys. Chem. Lett. 10 (2019) 3094-3100. DOI:10.1021/acs.jpclett.9b00842 |
| [20] |
Y.F. Huang, X. Zhou, R. Zhou, et al., Chem. Eur. J. 20 (2014) 5640-5648. DOI:10.1002/chem.201400011 |
| [21] |
Q. Xu, P. Pu, J.G. Zhao, et al., J. Mater. Chem. A: Mater. Energy Sustain. 3 (2015) 542-546. DOI:10.1039/C4TA05483K |
| [22] |
Z. Zhang, J.H. Hao, J. Zhang, B.L. Zhang, J.L. Tang, RSC Adv. 2 (2012) 8599-8601. DOI:10.1039/c2ra21217j |
| [23] |
J.C. Ge, Q.Y. Jia, W.M. Liu, et al., Adv. Mater. 27 (2015) 4169-4177. DOI:10.1002/adma.201500323 |
| [24] |
X.W. Hua, Y.W. Bao, H.Y. Wang, Z. Chen, F.G. Wu, Nanoscale 9 (2017) 2150-2161. DOI:10.1039/C6NR06558A |
| [25] |
K. Jiang, S. Sun, L. Zhang, et al., Angew. Chem. Int. Ed. 54 (2015) 5360-5363. DOI:10.1002/anie.201501193 |
| [26] |
Y.Z. Fan, Y. Zhang, N. Li, et al., Sens. Actuators B: Chem. 240 (2017) 949-955. DOI:10.1016/j.snb.2016.09.063 |
| [27] |
Z.G. Wang, B.S. Fu, S.W. Zou, et al., Nano Res. 9 (2016) 214-223. DOI:10.1007/s12274-016-0992-2 |
| [28] |
S.J. Zhu, Q.N. Meng, L. Wang, et al., Angew. Chem. Int. Ed. 52 (2013) 3953-3957. DOI:10.1002/anie.201300519 |
| [29] |
P.L. Li, S. Liu, W.W. Cao, et al., Chem. Commun. (Camb.) 56 (2020) 2316-2319. DOI:10.1039/C9CC09223D |
| [30] |
P.L. Gao, J.W. Wang, M. Zheng, Z.G. Xie, Chem. Eng. J. 381 (2020) 122665. DOI:10.1016/j.cej.2019.122665 |
| [31] |
Y.Z. Fu, S.L. Wu, H.K. Zhou, et al., Ind. Eng. Chem. Res. 59 (2020) 1723-1729. DOI:10.1021/acs.iecr.9b06289 |
| [32] |
H.H. Ran, X.T. Cheng, Y.W. Bao, et al., J. Mater. Chem. B: Mater. Biol. Med. 7 (2019) 5104-5114. DOI:10.1039/C9TB00681H |
| [33] |
X.Y. Teng, C.G. Ma, C.J. Ge, et al., J. Mater. Chem. B: Mater. Biol. Med. 2 (2014) 4631-4639. DOI:10.1039/c4tb00368c |
| [34] |
P.L. Li, X. Yang, X.H. Zhang, et al., J. Mater. Sci. 55 (2020) 16744-16757. DOI:10.1007/s10853-020-05262-6 |
| [35] |
G. Gao, Y.W. Jiang, J.J. Yang, F.G. Wu, Nanoscale 9 (2017) 18368-18378. DOI:10.1039/C7NR06764J |
| [36] |
X.W. Hua, Y.W. Bao, Z. Chen, F.G. Wu, Nanoscale 9 (2017) 10948-10960. DOI:10.1039/C7NR03658B |
| [37] |
J.J. Yang, G. Gao, X.D. Zhang, et al., Nanoscale 9 (2017) 15441-15452. DOI:10.1039/C7NR05613C |
| [38] |
X. Geng, Y.Q. Sun, Z.H. Li, et al., Small 15 (2019) e1901517. DOI:10.1002/smll.201901517 |
| [39] |
K.J. Jiang, S.Z. Hu, Y.C. Wang, Z.J. Li, H.W. Lin, Small 16 (2020) e2001909. DOI:10.1002/smll.202001909 |
| [40] |
W.Y. Lv, M. Lin, R.S. Li, et al., Chin. Chem. Lett. 30 (2019) 1410-1414. DOI:10.1016/j.cclet.2019.04.011 |
| [41] |
W.D. Li, Y. Liu, B.Y. Wang, et al., Chin. Chem. Lett. 30 (2019) 2323-2327. DOI:10.1016/j.cclet.2019.06.040 |
| [42] |
B.Y. Wang, J. Li, Z.Y. Tang, B. Yang, S.Y. Lu, Sci. Bull. (Beijing) 64 (2019) 1285-1292. DOI:10.1016/j.scib.2019.07.021 |
| [43] |
S.Y. Lu, L.Z. Sui, J.J. Liu, et al., Adv. Mater. 29 (2017) 1603443. DOI:10.1002/adma.201603443 |
| [44] |
B. Li, S. Zhao, L. Huang, et al., Chem. Eng. J. 408 (2021) 127245. DOI:10.1016/j.cej.2020.127245 |
| [45] |
X.C. Li, S.J. Zhao, B.L. Li, et al., Coord. Chem. Rev. 431 (2020) 213686. |
| [46] |
H.Q. Song, X.J. Liu, B.Y. Wang, Z.Y. Tang, S.Y. Lu, Sci. Bull. (Beijing) 64 (2019) 1788-1794. DOI:10.1016/j.scib.2019.10.006 |
| [47] |
H. Nie, M.J. Li, Q.S. Li, et al., Chem. Mater. 26 (2014) 3104-3112. DOI:10.1021/cm5003669 |
| [48] |
L.L. Pan, S. Sun, A.D. Zhang, et al., Adv. Mater. 27 (2015) 7782-7787. DOI:10.1002/adma.201503821 |
| [49] |
A. Sharma, T. Gadly, A. Gupta, et al., J. Phys. Chem. Lett. 7 (2016) 3695-3702. DOI:10.1021/acs.jpclett.6b01791 |
| [50] |
B. van Dam, H. Nie, B. Ju, et al., Small 13 (2017) 1702098. DOI:10.1002/smll.201702098 |
| [51] |
C. Settembre, A. Fraldi, D.L. Medina, A. Ballabio, Nat. Rev. Mol. Cell Biol. 14 (2013) 283-296. DOI:10.1038/nrm3565 |
| [52] |
X.D. Zhang, X.K. Chen, Y.X. Guo, et al., Nanoscale Horiz. 5 (2020) 481-487. DOI:10.1039/C9NH00643E |
| [53] |
X.K. Chen, X.D. Zhang, L.Y. Xia, et al., Nano Lett. 18 (2018) 1159-1167. DOI:10.1021/acs.nanolett.7b04700 |
| [54] |
X.K. Chen, X.D. Zhang, F.M. Lin, Y.X. Guo, F.G. Wu, Small 15 (2019) 1901647. DOI:10.1002/smll.201901647 |
| [55] |
X.K. Chen, X.D. Zhang, C.C. Li, et al., Sens. Actuators B: Chem. 295 (2019) 49-55. DOI:10.1016/j.snb.2019.05.031 |
| [56] |
X.D. Zhang, X.K. Chen, S.Q. Kai, et al., Anal. Chem. 87 (2015) 3360-3365. DOI:10.1021/ac504520g |
| [57] |
L.L. Wu, X.L. Li, Y.F. Ling, C.S. Huang, N.Q. Jia, ACS Appl. Mater. Interfaces 9 (2017) 28222-28232. DOI:10.1021/acsami.7b08148 |
| [58] |
D.Y. Zhang, Y. Zheng, H. Zhang, et al., Nanoscale 9 (2017) 18966-18976. DOI:10.1039/C7NR05349E |
| [59] |
Y.Y. He, Z.X. Li, Q.Y. Jia, et al., Chin. Chem. Lett. 28 (2017) 1969-1974. DOI:10.1016/j.cclet.2017.07.027 |
| [60] |
E. Shuang, Q.X. Mao, X.L. Yuan, et al., Nanoscale 10 (2018) 12788-12796. DOI:10.1039/C8NR03453B |
| [61] |
Q.Q. Zhang, T. Yang, R.S. Li, et al., Nanoscale 10 (2018) 14705-14711. DOI:10.1039/C8NR03212B |
| [62] |
H.F. Liu, Y.Q. Sun, Z.H. Li, et al., Nanoscale 11 (2019) 8458-8463. DOI:10.1039/C9NR01678C |
| [63] |
S. Chen, Y. Jia, G.Y. Zou, Y.L. Yu, J.H. Wang, Nanoscale 11 (2019) 6377-6383. DOI:10.1039/C9NR00039A |
| [64] |
S.J. Zhao, S.L. Wu, Q.Y. Jia, et al., Chem. Eng. J. 388 (2020) 124212. DOI:10.1016/j.cej.2020.124212 |
| [65] |
H.Y. Qin, Y.Q. Sun, X. Geng, et al., Anal. Chim. Acta 1106 (2020) 207-215. DOI:10.1016/j.aca.2020.02.002 |
| [66] |
H. Singh, S. Sreedharan, K. Tiwari, et al., Chem. Commun. 55 (2019) 521-524. DOI:10.1039/C8CC08610A |
| [67] |
S. Guo, Y.Q. Sun, X. Geng, et al., J. Mater. Chem. B: Mater. Biol. Med. 8 (2020) 736-742. DOI:10.1039/C9TB02043H |
| [68] |
L.L. Tong, X.X. Wang, Z.Z. Chen, et al., Anal. Chem. 92 (2020) 6430-6436. DOI:10.1021/acs.analchem.9b05553 |
| [69] |
S. Hirayama, K. Shobatake, K. Tabayashi, Chem. Phys. Lett. 121 (1985) 228-232. DOI:10.1016/0009-2614(85)85516-0 |
| [70] |
A. Gorman, J. Killoran, C. O'Shea, J. Am. Chem. Soc. 126 (2004) 10619-10631. DOI:10.1021/ja047649e |
 2021, Vol. 32
2021, Vol. 32 

