b State Key Laboratory of Virology, Wuhan Institute of Virology, Center for Biosafety Mega-Science, Chinese Academy of Sciences (CAS), Wuhan 430071, China;
c State Key Laboratory of Virology, College of Life Sciences, Wuhan University, Wuhan 430072, China
The outbreak of a coronavirus-associated acute respiratory disease in December 2019, which are now called coronavirus disease 19 (COVID-19), has rapidly affected the entire world in a few months [1, 2]. By far, more than 105, 000, 000 cases were confirmed, with more than 2, 300, 000 deaths. The pathogen has been identified as a human and bat severe acute respiratory syndrome coronaviruses (SARS-CoVs) and was designated as SARS-CoV-2 [3]. Nowadays, there are no specific pharmacological treatments available to treat SARS-CoV-2 patients [4]. Most of the patients were treated by broad-spectrum antiviral drugs, such as Remdesivir and Lopinavir [5], or immunomodulatory and anti-inflammatory drugs, such as Barictinib [6]. Chloroquine (CQ), a medication primarily used to treat malaria for over 350 years [7], received great attention for the potential treatment of COVID-19 during the early days of the outbreak, as in vitro study demonstrated CQ was able to inhibit the growth of SARS-CoV-2 [2, 5]. CQ can block the infection of virus by increasing the endosomal pH, whereas a lower pH is required for virus fusion. CQ was also found to interfere with the terminal glycosylation of cellular receptors of SARS-CoV [8]. In addition, CQ was previously reported as a broad-spectrum antiviral drug, against HIV/AIDS and H5N1 virus [9, 10]. In March 2020, CQ was designated by the FDA as "the emergency use authorization (EUA)" medicine to treat certain hospitalized patients with COVID-19 when a clinical trial was unavailable, or participation in a clinical trial was not feasible. Later the FDA revoked the EUA status of CQ due to serious cardiac adverse events and other potential serious side effects including liver toxicity [11]. Therefore, it is of great value to develop safe and more effective formulations of CQ to treat COVID-19 at high concentrations.
Cucurbit[7]uril (CB[7]), as a family member of cucurbit[n]urils (CB[n], n = 5–8, 10, 13–15) macrocycles, has been widely used for various biomedical applications due to its good water solubility and biocompatibility, and high binding affinity towards bioactive molecules in aqueous solutions [12-15]. In recent years, CB[7]-based supramolecular formulations through direct host-guest encapsulation strategy have been well demonstrated to reduce the non-specific toxicity of the guest drugs [14-16]. For instance, Zhang and co-workers developed CB[7]-based supramolecular formulations of anticancer agents for the targeted release of the guest drugs, to reduce the side-effects and improve the therapeutic efficacy during cancer chemotherapy [17, 18]. Indeed, the overexpressed spermine (SP) inside certain types of cancer cells can activate the supramolecular formulations through the competitive replacement induced release of guest chemotherapeutics from CB[7]'s cavity, as CB[7] exhibits a much higher binding affinity with SP than that with guest drugs [18]. SP belongs to polyamine family which is found to involve in many RNA life cycles, including replication of RNA viruses, viral protein translation, RNA polymerase activity keeping, and RNA infectivity maintaining [19-21]. The depletion of polyamines by introducing inhibitors of polyamine synthases has been considered as a promising approach in restricting the RNA virus infection. Therefore, we hypothesized that free CB[7] may exhibit antiviral activity due to the supramolecular sequestration of SP molecules. In addition, the supramolecular formulation of CQ by CB[7] may reduce its non-specific toxicity, and improve the overall antiviral activity due to the synergistic actions of CB[7] and CQ via different mechanisms (Fig. 1).

|
Download:
|
| Fig. 1. Schematic illustration of the design of CQ-CB[7] supramolecular formulation for coronavirus treatment. | |
Herein, we employed CB[7] to encapsulate CQ to afford a novel supramolecular formulation of CQ, which could reduce the non-specific toxicity of CQ due to the supramolecular protection that would minimize the non-specific interactions of CQ with non-target proteins. CQ could be released from the cavity of CB[7] upon the competitive binding of the polyamines inside the cells, to exert its antiviral activity. Meanwhile, the sequestration of polyamine by CB[7] would likely disrupt the interactions of polyamine with viral RNA, which is otherwise required for RNA replication, thus providing a secondary antiviral action. This study may revive CQ for the treatment of COVID-19 with a safer and more effective supramolecular formulation.
First, the successful formation of host-guest complexes between CQ and CB[7] was investigated.1H NMR spectroscopic titration was performed to understand the binding sites of CQ-CB[7] host-guest complexes. As shown in Fig. 2A, the proton resonances of CQ exhibited up-field and down-field shifts, upon addition of CB[7]. In particular, the protons in the aliphatic chain (H6, H7, H8, H9) and in the aromatic ring (H2, H3, H4) exhibited an upfield shift, indicating that both these two sites can be included inside the hydrophobic cavity of CB[7]. Considering the repulsion effects if two CB[7] are respectively sitting over these two sites, CB[7] may shuttle between the two sites frequently.
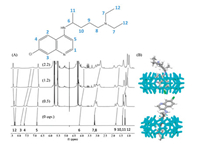
|
Download:
|
| Fig. 2. (A) The 1H NMR spectra of CQ in the absence (bottom), and presence of 0.5, 1.2 and 2.2 equiv. CB[7]. The protons of CQ were numerically labelled, and dash lines represent the protons shift; (B) Energy-minimized three-dimensional chemical models of CQ-CB[7] complexes (MM2). | |
With the assistance of MM2 calculations, the energy-minimized three-dimensional chemical model of the two possible CQ-CB[7] host-guest complexes were also obtained, as shown in Fig. 2B, supporting two possible binding modes (or two individual binding stages of the molecular shuttle).
Subsequently, the binding stoichiometry of CQ-CB[7] host-guest complexes was assessed by Job plot titration and ESI-MS analysis. The Job plot of CQ and CB[7] at 330 nm derived from UV–vis continuous variation titration demonstrated a maximum ΔA at 0.5 of [CQ]/([CQ]+[CB[7]]), indicating a 1:1 binding ratio between CQ and CB[7] (Fig. 3A). In addition, the main doubly charged peak at m/z 741.7703 was found in the ESI-MS spectrum, consistent with the calculated m/z value of 741.7704 for [CQ-CB[7]+2H]2+, further confirming the 1:1 binding stoichiometry of CQ-CB[7] complexes (Fig. 3B).

|
Download:
|
| Fig. 3. (A) UV–vis absorbance spectra of CQ titrated by CB[7] (inset: the Job plot ΔA at 330 nm); (B) ESI-MS spectrum of CQ-CB[7], a doubly charge peak found at m/z 741.7703, corresponding to [CQ-CB[7]+2H]2+; (C) The dependence of ΔH against molar ratios between CQ and CB[7], fitted by using "one set of binding sites" binding model; (D) Thermogram of 19 drops of CB[7] (2.0 mmol/L) titrated into CQ (0.1 mmol/L) during ITC titration. | |
Finally, the binding behaviours of CQ-CB[7] host-guest complexes were further investigated by isothermal titration calorimetry (ITC), to understand the binding affinity, binding stoichiometry as well as the thermodynamic parameters during the binding process. As shown in Figs. 3C and D, by using "one set of binding sites" binding model to fit the dependence of ΔH against the molar ratios between CQ and CB[7], a 1:1 binding stoichiometry of CQ-CB[7] host-guest complexes (N sites = 1.19) was obtained, with a binding affinity Ka of 4.24±0.73×104 L/mol, thermodynamically driven by both enthalpy change (ΔH = -16.7 kJ/mol) and the entropy change (TΔS = 9.8 kJ/mol). The enthalpy and entropy changes during the binding process were driven by the coulombic attractions, hydrogen bonding, hydrophobic effects and the release of water from CB[7]'s cavity upon inclusion [22, 23].
Before evaluating the cytotoxicity of supramolecular formulation of CQ-CB[7] complexes, the in vitro biocompatibility of CQ and CB[7] was individually studied by using Vero E6 (African green monkey kidney epithelial cells) and L-02 (human health liver cells), respectively. As shown in Figs. 4A and B, after incubating cells with CQ for 48 h, a dose-dependent cytotoxicity was observed in both Vero E6 and L-02 cell lines, with the IC50 values of 34.88 μmol/L and 12.64 μmol/L, respectively determined. The lower IC50 value of L-02 than that of Vero E6 indicated a possibly higher hepatotoxicity of CQ than its nephrotoxicity [24]. In contrast, CB[7] showed relatively good in vitro biocompatibility towards both cell lines at up to millimolar concentration range, consistent with the previous studies (Figs. 4C and D) [25, 26].

|
Download:
|
| Fig. 4. (A-D) Cytotoxicity of different concentrations of CQ and CB[7] molecules; (E, F) 60 μmol/L CQ co-treated with different concentrations of CB[7] (0-600 μmol/L); towards Vero E6 and L-02 cell lines after incubation for 48 h. Data were presented as mean±SD, by One-way ANOVA followed by Dunnett's test, n ≥ 5 independent experiments. | |
To obtain the optimal amount of CB[7] in the supramolecular formulation of CQ-CB[7], a constant concentration of CQ (60 μmol/L) with different equivalents of CB[7] (ranging from 0.5 to 10) were used to treat Vero E6 and L-02 cell lines for 48 h. As shown in Figs. 4E and F, the co-treatment of CQ with different equivalents of CB[7] exhibited an improved cell viability, with a CB[7] dose-dependency. Of a significant note, the cytotoxicity of CQ (60 μmol/L) against both Vero E6 and L-02 cell lines was completely inhibited in the presence of 300 μmol/L and 600 μmol/L CB[7], respectively. Considering the binding constant of CQ-CB[7] complexes (~104 L/mol) and the IC50 value of CQ towards the two cell lines, to ensure the complexation, 300 μmol/L CB[7] was selected to prepare CQ-CB[7] supramolecular formulation for further study.
We assessed the antiviral activities of CQ-CB[7] formulation against SARS-CoV-2. As shown in Fig. 5A, CQ showed an inhibition effect dose-dependently in the SARS-CoV-2 RNA accumulation with a half-maximal effective concentration (EC50) value of 1.91 μmol/L in Vero E6 cells. In Fig. 5B, after encapsulated by CB[7] to from a CQ-CB[7] supramolecular formulation with 2 μmol/L CQ, the inhibition (%) of viral RNA replication were improved significantly. Notably, CB[7] alone was found to exhibit the remarkable inhibition effect towards the virus replication, which might be attributed to the inhibition of polyamine synthesis inside the cells by CB[7] that resulted in the disturbance of the virus replication [27].

|
Download:
|
| Fig. 5. (A, B) The inhibition (%) of CQ and CQ-CB[7] formulation on the SARS-CoV-2 RNA accumulation in Vero E6 cells; and (C, D) the inhibition (%) of CQ and CQ-CB[7] formulation on MHV-A59 RNA accumulation in N2A cells. Data were presented as mean±SD, by One-way ANOVA followed by Dunnett's test, n ≥ 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 versus the corresponding with infection group. | |
Moreover, we also assessed the antiviral activities of CQ-CB[7] formulation against another coronavirus MHV-A59. As shown in Fig. 5C, CQ showed an inhibition effect dose-dependently in the MHV-A59 replication with EC50 value of 1.65 μmol/L in N2A cells. In Fig. 5D, after encapsulated by CB[7] to from a CQ-CB[7] supramolecular formulation with 0.5 μmol/L or 1 μmol/L CQ, the inhibition (%) of viral RNA replication was also improved significantly. Again, CB[7] alone was found to exhibit the remarkable inhibition effect towards the MHV-A59 replication in N2A cell line.
In summary, this work demonstrated that the supramolecular formulation of CQ-CB[7] could reduce the non-specific hepatotoxicity and nephrotoxicity of CQ and achieve host-guest synergistic antiviral activity towards both SARS-CoV-2 and MHV-A59 in vitro. CQ exerts its antiviral activity upon release from the cavity of CB[7], induced by polyamines' competitive displacement. Meanwhile, the supramolecular complexation of polyamine by CB[7] disrupts the interactions of polyamine with viral RNA, which is otherwise required for RNA replication. This is the first report about the anti-SARS-CoV-2 activity of a synthetic receptor. And this study provides a safer and more effective formulation of CQ for anti-SARS-CoV-2 action, and may revive CQ for clinical treatment of COVID-19 patients upon further development.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentsWe acknowledge the National Virus Resource Center for the viral resource support. This work was supported by the Science and Technology Development Fund, Macau SAR (No. 0007/2020/A), International Partnership Program of Chinese Academy of Sciences (No. 153B42KYSB20200004 to X. Zhou and R. Wang), the Strategic Priority Research Program of CAS (No. XDB29010300), and the National Natural Science Foundation of China (Nos. 21871301, 22071275, 31970169, 31800140, 31800140 and 31670161), the Yunde Hou Academician Fund from National Institute for Viral Disease Control and Prevention (No. 2019HYDQNJJ10).
Appendix A. Supplementary dataSupplementary material related to this article can be found, in the online version, at doi: https://doi.org/10.1016/j.cclet.2021.04.008.
| [1] |
X. Yang, Y. Yu, J. Xu, et al., Lancet Respir. Med. 8 (2020) 475-481. DOI:10.1016/S2213-2600(20)30079-5 |
| [2] |
A. Cortegiani, G. Ingoglia, M. Ippolito, et al., J. Crit. Care 57 (2020) 279-283. DOI:10.1016/j.jcrc.2020.03.005 |
| [3] |
A.E. Gorbalenya, S.C. Baker, R.S. Baric, et al., Nat. Microbiol. 5 (2020) 536-544. DOI:10.1038/s41564-020-0695-z |
| [4] |
T. Shu, M. Huang, D. Wu, et al., Virol. Sin. 35 (2020) 321-329. DOI:10.1007/s12250-020-00242-1 |
| [5] |
M. Wang, R. Cao, L. Zhang, et al., Cell Res. 30 (2020) 269-271. DOI:10.1038/s41422-020-0282-0 |
| [6] |
E.G. Favalli, M. Biggioggero, G. Maioli, et al., Lancet Infect. Dis. 20 (2020) 1012-1013. |
| [7] |
Andrade-Neto V.F., M.G.L. Brandão, J.R. Stehmann, et al., J. Ethnopharmacol. 87 (2003) 253-256. DOI:10.1016/S0378-8741(03)00141-7 |
| [8] |
M.J. Vincent, E. Bergeron, S. Benjannet, et al., Virol. J. 2 (2005) 69. DOI:10.1186/1743-422X-2-69 |
| [9] |
A. Savarino, Di Trani L., I. Donatelli, et al., Lancet Infect. Dis. 6 (2006) 67-69. DOI:10.1016/S1473-3099(06)70361-9 |
| [10] |
Y. Yan, Z. Zou, Y. Sun, et al., Cell Res. 23 (2013) 300-302. DOI:10.1038/cr.2012.165 |
| [11] |
M.G.S. Borba, F.F.A. Val, V.S. Sampaio, et al., JAMA Network Open 3 (2020) e208857. DOI:10.1001/jamanetworkopen.2020.8857 |
| [12] |
S.J. Barrow, S. Kasera, M.J. Rowland, et al., Chem. Rev. 115 (2015) 12320-12406. DOI:10.1021/acs.chemrev.5b00341 |
| [13] |
D. Shetty, J.K. Khedkar, K.M. Park, et al., Chem. Soc. Rev. 44 (2015) 8747-8761. DOI:10.1039/C5CS00631G |
| [14] |
K.I. Kuok, S. Li, I.W. Wyman, et al., Ann. N. Y. Acad. Sci. 1398 (2017) 108-119. DOI:10.1111/nyas.13376 |
| [15] |
H. Yin, R. Wang, Israel J. Chem. 58 (2018) 188-198. DOI:10.1002/ijch.201700092 |
| [16] |
H. Yin, X. Zhang, J. Wei, et al., Theranostics 11 (2021) 513-1526. |
| [17] |
Y. Chen, Z. Huang, J.F. Xu, et al., ACS Appl. Mater. Interfaces 8 (2016) 22780-22784. DOI:10.1021/acsami.6b08295 |
| [18] |
Y. Chen, Z. Huang, H. Zhao, et al., ACS Appl. Mater. Inter. 9 (2017) 8602-8608. DOI:10.1021/acsami.7b01157 |
| [19] |
V. Mastrodomenico, J.J. Esin, M.L. Graham, et al., J. Virol. 93 (2019) e00530-00519. |
| [20] |
T.M. Kicmal, P.M. Tate, C.N. Dial, et al., J. Virol. 93 (2019) e01054-01019. |
| [21] |
B.C. Mounce, E.Z. Poirier, G. Passoni, et al., Cell Host Microbe 20 (2016) 167-177. DOI:10.1016/j.chom.2016.06.011 |
| [22] |
Y. Jang, R. Natarajan, Y. Ko, et al., Angew. Chem. Int. Ed. 53 (2014) 1003-1007. DOI:10.1002/anie.201308879 |
| [23] |
F. Biedermann, V. Uzunova, O. Scherman, et al., J. Am. Chem. Soc. 134 (2012) 15318-15323. DOI:10.1021/ja303309e |
| [24] |
M. Shichiri, N. Kono, Y. Shimanaka, et al., J. Biol. Chem. 287 (2012) 2926-2934. DOI:10.1074/jbc.M111.321281 |
| [25] |
H. Chen, J.Y.W. Chan, X. Yang, et al., RSC Adv. 5 (2015) 30067-30074. DOI:10.1039/C5RA04335B |
| [26] |
X. Zhang, X. Xu, S. Li, et al., Sci. Rep. 8 (2018) 8819. DOI:10.1038/s41598-018-27206-6 |
| [27] |
C. Parente Carvalho, A. Norouzy, V. Ribeiro, et al., Org. Biomol. Chem. 13 (2015) 2866-2869. DOI:10.1039/C4OB02122C |
 2021, Vol. 32
2021, Vol. 32 

