In recent years, cell membrane-camouflaged drug delivery systems (CMCDDs) have attracted significant research attention. They assembles nanoparticles with cell membrane materials to completely retain the original protein expression and structural functions of the cell membrane. Additionally, the synthetic chemical, core nanoparticles support the cell membrane structure while loading the drug. Numerous studies have confirmed that cell membrane materials improve the biocompatibility of drug delivery systems and enhance the targeting ability of target tissues and cells [1-4]. In particular, erythrocyte membrane-camouflaged nanoparticles exhibit long circulation performance in vivo [5], and macrophage membrane and tumor cell membrane-camouflaged nanoparticles could can be efficiently targeted with target inflammatory factors and tumors [6, 7]. These CMCDDSs have shown a promising potential in the field of drug delivery systems and have a vast scope of application.
A large number of preclinical studies have found that CMCDDSs have significant therapeutic effects in animal models. Moreover, a variety of cell membrane biomimetic preparations, developed by Cellics US Inc., have entered phase 1b/2a clinical trials. With the development of cell membrane camouflaged drug delivery systems, there has been an increasing number of related patent applications and proof of concept (POC) clinical researches in the recent years. We list the representational patent for the CMCDDSs (Table 1), and the proof of concept (POC) clinical applications and researches of the CDCDDs (Table 2). However, a host of CMCDDS studies have found that, while cell membrane materials provide powerful biological functions, their stability after extraction can also limit the development of CMCDDSs. First, challenges exist in the CMCDDS preparation process, including in cell membrane material extraction, oriented assembly, and continuous in-scale production. Second, CMCDDSs must be provided with a variety of in vivo biological functions to better adapt to the complex pathological environment. Third, CMCDDS studies must be combined with personalized treatment, to further promote their precise clinical application. Therefore, herein, we provide an overview of these challenges and the corresponding solution strategies. Additionally, we analyze the frontier applications of CMCDDSs in medicine, such as in anti-inflammatory applications, anti-pathogenic microbial, and bio-detoxification.
|
|
Table 1 Representational patent for the CMCDDSs. |
|
|
Table 2 The POC clinical applications and researches of the CMCDDSs. |
The extraction and oriented assembly of cell membrane materials is a prerequisite for the successful preparation of CMCDDSs. It is necessary to improve the extraction rate of cell membrane materials and enhance the oriented assembly of cell membrane materials using core nanoparticles. This ensures that the relevant functional proteins are correctly exposed and perform their corresponding biological functions.
2.1. Refinement of the cell membrane material extraction processThe extraction of erythrocyte membranes is relatively simple compared to the extraction of other cell membrane materials because the techniques and equipment used for blood product preparation provide a mature basis for red blood cell (RBC) membrane extraction. Due to the nucleus-free structure of the erythrocytes, the purity of erythrocyte membrane material extraction will is higher than that compared of other cell membranes.
Therefore, further improving the extraction purity of nucleated cell membrane materials is an important future research direction of cell membrane extraction. Large volume extraction of cell membrane materials is a prerequisite for the clinical application of CMCDDSs. For the extraction of cell membranes from different cells, either density gradient centrifugation or differential centrifugation is generally used. However, the choice of cell fragmentation methods and centrifugation parameter conditions has not been clearly demonstrated. More importantly, after the membrane material is extracted and separated, their stability decreases sharply because of the lack of cytoplasmic support and nutrient supply. Studies have shown that the platelet membrane can only be stored at 22–24 ℃ for 5–7 days [8], which undoubtedly increases the risk of bacterial infection. Cell factories, which are fully automated cell culture systems, are a proven solution for the large-scale production of cells, vaccines, and therapeutic proteins in systems that have the same growth kinetics as laboratory-scale, cell culture products [9]. Therefore, cell factories are a current solution for the problem of cell membrane material sources. We suggest that future studies should optimize existing cell membrane extraction techniques alongside the design of extraction devices that can be combined with cell factory technology to establish an assembly line for cell culture and cell membrane extraction. As for the cell membrane purity, suggests cell sorting technology and protein affinity purification–mass spectrometry to analyze the unique antigens in the cell membrane materials to achieve purification and separation of the extracted products [10]. We believe that with the promotion of CMCDDS transforming clinical technologies, as potential problems are identified, the development of CMCDDSs will be continuously advanced.
2.2. Promoting the oriented assembly of CMCDDSsThe assembly of CMCDDSs is attributed to the charge asymmetry of cell membranes, resulting in the spontaneous formation of a core–shell nanostructure [11]. A number of early studies have promoted the directional assembly of CMCDDSs through physical methods, mainly ultrasound [12-14], electric shock [15, 16], extrusion [17, 18], and freeze-thaw [19]. A number of studies found that the success rate of the above methods was only 15% [12]. The success rate of cell membrane-directed assembly decreases with an increase in the number of cell membrane processing steps before assembly. Therefore, simplifying the cell membrane assembly steps is important for efficient assembly.
An important influencing factor that affects the biological function of CMCDDSs is the correct exposure of the functional protein on one side of the cell membrane. A number of studies have confirmed that erythrocyte membrane-camouflaged nanoparticles only that 84% of the cell membranes are assembled in the correct orientation [20]. We used the erythrocyte membrane transmembrane protein, Band3, as the assembly "handler", and designed a peptide ligand, P4.2 (LFVRRGQPFTIILYF), with high affinity to the intracellular domain of the Band3 protein. The P4.2 peptide was modified on the surface of the core liposome to direct the oriented assembly of the cell membrane to the core carrier through the specific binding between the intracellular section of the Band3 protein and P4.2 peptide [21]. This strategy provides a new approach for the oriented assembly of CMCDDSs (Fig. 1).

|
Download:
|
| Fig. 1. Summary of the targeted fabrication of RCB membrane -coated liposomes (RBC-LIP). The P4.2-derived peptide ligand was modified on AmB-loaded liposomes using a postinsertion technique. Reproduced with permission [21]. Copyright 2019, American Chemical Society. | |
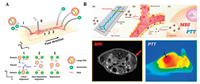
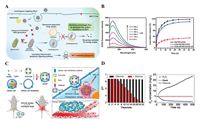
Stable and continuous industrial production is an important factor in the future development of CMCDDSs. Continuous preparation of CMCDDSs utilizing microfluidics has attracted significant research attention. Liu et al. utilized microfluidics to separate the cell membrane material from the cell culture fluid, which can be used for the continuous preparation of CMCDDSs [22]. This method can achieve high separation purity (> 90%) and recovery (> 80%) of cell membrane materials (Fig. 2A). Rao et al. also used microfluidics, combined with electroporation methods, which can efficiently wrap Fe3O4 nanoparticles inside the cell membranes [23].In vivo studies have shown that RBC-MNs were used for enhanced tumor magnetic resonance image (MRI) and photothermal therapy (PTT) (Fig. 2B). Compared with the traditional squeeze method, the application of microfluidics to structure CMCDDSs can reduce the preparation expense and satisfy the requirements for stable and continuous production.
|
Download:
|
| Fig. 2. (A) Schematic of the microfluidic chip for exosome separation from large EV. Reproduced with permission [22]. Copyright 2017, American Chemical Society. (B) Microfluidic electroporation-facilitated synthesis of erythrocyte membrane-capped magnetic nanoparticles (RBC-MNs) for enhanced imaging-guided cancer therapy. Reproduced with permission [23]. Copyright 2017, American Chemical Society. | |
With in-depth research on CMCDDSs, an increasing number of studies have found that the biological functions provided solely by the cell membranes are not suitable for the complex physiological environment of the pathogenic site and for satisfying potential therapy requirements. Solely relying on the biological functions of the cell membrane itself cannot satisfy the requirements for multi-functional CMCDDSs. Therefore, the multi-functional design of CMCDDSs requires significant research attention.
3.1. Application of diversified cell membrane materialsRelative to the different purposes of drug delivery, the cell membrane materials in CMCDDSs can include erythrocyte membranes [24, 25], leukocyte membranes [26], cancer cell membranes [27], and platelet membranes [28, 29], because different cell membrane materials provide different biological functions. Through the fusion of multiple eukaryotic cell membrane materials to create multi-cell membrane fusion materials, CMCDDSs can be simultaneously ingrained with the biological functions of multiple cell membranes expanding the sources of cell membrane materials, the discovery of novel cell membrane materials can provide new directions for CMCDDS research. Surface modification of cell membrane materials is an important strategy for the multi-functional construction of CMCDDSs, allowing them to not only inherits the biological functions of the natural cell membrane, but also can obtain an "artificial" new function as designed.
3.1.1. Multicellular membrane fusion strategyDue to the fluidity of cell membranes, fusion of different types of cell membrane materials can occur to create multi-cell membrane fusion materials [30]. Han et al. fused tumor cell and RBC membranes to gain a multi-cell membrane-camouflaged drug delivery system (nanoAg@erythrosome). Since this drug delivery system had the antigens of two cell membrane materials, the combination of αPD-L1 treatments effectively inhibited tumor proliferation [31]. Jiang et al. fused RBC and MCF-7 cell membranes and coated the product with melanin nanoparticles to structure Melanin@RBC-M. This CMCDDS showed both excellent long circulation and tumor-specific targeting in vivo, and exhibited significantly enhanced PTT effects [32]. This strategy is a method for fusing two eukaryotic cell membranes to form a novel, fused cell membrane material, which has been extensively researched for use in tumor cell membranes and leukocyte membranes [33], RBC membranes and platelet membranes [34], and fusion membranes of different apoptotic bodies [35]. In addition, Chen et al. fused eukaryotic tumor cell membrane vesicles (CMVs) and prokaryotic Salmonella outer membrane vesicles (OMVs) to obtain a novel, fused, cell membrane material (EPV), which was assembled with PLGA-ICG (PI) to form PI@EPV [36]. In this CMCDDS, the CMVs realized the specific targeting of PI@EPV to the cancer cell, and the OMVs enhanced the immune response of the tumor (Fig. 3).
|
Download:
|
| Fig. 3. Schematic illustration of fabrication of eukaryotic–prokaryotic vesicles coated PI@EPV nanovaccine. Reproduced with permission [36]. Copyright 2020, Wiley Publishing Group. | |
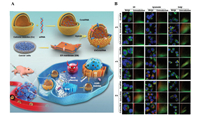
In recent years, the selection of cell membrane materials has mainly focused on the outer membrane of eukaryotic cells in the majority of studies. CMCDDSs structured from cell membranes, inner membranes, or prokaryotic cell membranes, as a novel type of CMCDDS, have also attracted attention. Qiu et al. extracted the endoplasmic reticulum (ER) membrane to modify the siRNA delivery system (Fig. 4). This CMCDDS effectively avoids lysosomal enzymatic digestion, through the mediation of various protein signals, such as SNAREs and resident proteins on the ER membrane [37]. The intracellular membrane system, as the largest intracellular endomembrane, controls a wide range of intracellular biological functions. Intracellular membrane-camouflaged drug delivery systems have significant research value. Gao et al. coated PLGANP nanoparticles with an extracellular vesicle membrane from S. aureus to prepare NP@EV, which was internalized by S. aureus-infected macrophages at a high efficiency [38]. This EV membrane-camouflaged drug delivery system effectively adsorbs deeply infected tissue in the body, allowing the drug to be delivered to the site of infection.
|
Download:
|
| Fig. 4. (A) The assembly process and delivery of ER membrane (EM)-hybrid small interfering RNA (siRNA)-loaded nanoplexes (EhCv/siRNA NPs). (B) Representative colocalization images of different NPs in endoplasmic reticulum (ER), lysosomes, and Golgi apparatus at different times detected by confocal laser scanning microscope. Reproduced with permission [37]. Copyright 2019, Nature Publishing Group. | |
Functional modification of the cell membrane material surface has been essential way of multi-functionalizing CMCDDSs in recent years. This technique involves utilizing the active chemical structure of functional, small molecules, peptides, and antibody proteins and modifying them by direct or indirect coupling with cell membrane materials. This strategy adds to the strong, in vivo, biological performance of cell membranes and further enhances their ability to target disease, promote immune regulation, and perform bioimaging.
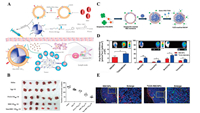
Functional small molecule modification strategies: Studies have modified folic acid (FA) to the surface of RBC membranes and coated the product with upconversion nanoparticles (UCNPs) to construct the RBS-UCNPs delivery system [39], which combines the significant advantages of long fluorescence lifetime, low toxicity, large penetration depth, and low damage to biological tissues. The RBS-UCNPs use tumor cells to overexpress FA receptors to achieve deep imaging. Ding et al. modified both FA and triphenylphosphine (TPP) on the surface of erythrocyte membranes to encapsulate UCNPs. This CMCDDS can achieve dual targeting of tumor cells and intracellular mitochondria. Under laser irradiation, the TPP can produce large amounts of reactive oxygen species in vivo, which disrupts the balance of mitochondrial reactive oxygen species, leading to mitochondrial dysfunction and tumor cell apoptosis [40]. Guo et al. constructed a mannitol-modified, erythrocyte membrane drug delivery system, which utilized the natural immune properties of mannitol (Figs. 5A and B). This mannitol-modified, erythrocyte membrane-camouflaged drug delivery system could efficiently bind to dendritic cells and enhance IFN-γ secretion and CD8+ T cell response [41].
|
Download:
|
| Fig. 5. (A) Schematic illustration of the preparation of Man-RBC membrane-coated PLGA-SShgp100 nanoparticles (Man-RBC-NPhgp) and induction of anti-tumor immunity. (B) In vivo tumor inhibition assays were performed after harvesting lungs for imaging and counting of tumor nodules larger than 2 mm in diameter manually. Reproduced with permission [41]. Copyright 2015, American Chemical Society. (C) Schematic of the preparation of DCDX-RBCNPs. Streptavidin-PEG-DSPE is synthesized and then inserted into RBC membranes. (D) In vivo distribution of RBCNPs and DCDX-RBCNPs in the brain. (E) The distribution of nanoparticles in the brain of tumor-bearing mice 14 days post-implantation. Reproduced with permission [44]. Copyright 2017, Elsevier Ltd. | |
Peptide and antibody protein modification strategies: Protein-protein interactions exist in every cell [42, 43]. Therefore, studies have modified cell membrane surfaces with peptides and antibody proteins to improve the ability to penetrate of complex biological barriers and target pathogenic microenvironments in CMCDDSs. Chai et al. modified the DCDX peptide (G DR DE DI DR DTG DR DA DE DR DW DS DE DK DF) on the surface of an RBC membrane (Figs. 5C–E). This promoted the ability of RBC membrane- camouflaged drug delivery systems to penetrate the blood brain barrier (BBB), and improve the therapeutic effect of U87 glioma [44]. Subsequently, this team modified the c(RGDyK) peptide on the surface of an RBC membrane, to achieve specific targeting of the erythrocyte membrane- camouflaged drug delivery system to tumors [45].
In addition to modifying peptides, modification of the cell membrane surface with antibody proteins has attracted substantial attention. The anti-EpCam was modified on RBC membranes by coating PAuNs nanoparticles to specifically target the EpCam receptor proteins on tumor cell membranes [46]. Studies have modified the surface of the RBC membrane with hyaluronidase (PH20), which not only extend the circulation time of the drug delivery system in vivo, but also utilizes PH20 to hydrolyze hyaluronan at the tumor, increasing rates of drug diffusion [47]. Dong et al. modified the TGF-β receptor II antibody (TGFβRII) on the leukemia cell membrane. This leukemia cell membrane-camouflaged drug delivery system efficiently targets the bone marrow microenvironment through the CXCR4 protein on the membrane surface of leukemic cells, and the release of αTGFβRII can effectively overcomes the drug resistance of the bone marrow microenvironment, significantly extending the survival of mice [48].
3.1.4. Bioengineering of cell membrane materials strategiesUtilizing cell biology techniques, cells can be pre-treated with gene editing or in vitro stimuli induced to express extra functional proteins on the cell membrane surface. This cell membrane surface engineering strategy, combined with cell biology technology, promotes the diversified development of cell membrane materials and expands the applications of CMCDDSs.
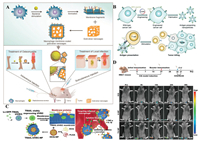
In vitro stimuli induction: Before cell membrane material extraction, studies focused on inducing cells to express functional proteins and promote drug loading through co-incubation with pathogenic microorganisms or chemotherapeutic drugs. This strategy has the advantage of simple operation. After co-incubation of macrophages with S. aureus and Escherichia coli bacteria, Wang et al. observed an increase in the expression of lipoprotein receptors (TLR1, TLR2, and TLR6) on the surface of macrophage membranes (Fig. 6A). The researchers extracted the above-treated macrophage membrane to coat with a gold-silver nanocage (GSNC), and prepared a TU-NPs cell membrane-camouflaged system, combined with the PTT function provided by GSNC. This CMCDDS significantly inhibited bacterial growth [49].
|
Download:
|
| Fig. 6. (A) The fabrication and application of Sa-M-GSNCs. A) Sa-M-GSNCs were fabricated through coating S. aureus pretreated macrophage membranes onto GSNCs. Reproduced with permission [49]. Copyright 2018, Wiley Publishing Group. (B) Wild-type cancer cells, which naturally present their own antigens via MHC-I, are engineered to express CD80, a co-stimulatory signal. The plasma membrane from these cells is then derived and coated onto polymeric nanoparticle cores. Reproduced with permission [53]. Copyright 2020, Wiley Publishing Group. (C) Schematic illustration of TU-NPs designed for targeting RA. (D)In vivo imaging results of ICG, U-NPs and TU-NPs after intravenous injection at the indicated time, n=3. Reproduced with permission [55]. Copyright 2020, Elsevier Ltd. | |
Unlike the induction chemotherapeutic drugs are co-incubated directly with the cells to obtain drug-loaded, cell membrane exosomes [50, 51]. Recently, studies have used curcumin to induce macrophages where the macrophage cells produce curcumin-loaded exosomes. This CMCDDS effectively improves the entrapment efficacy of curcumin. The study utilized the expression of the LFA-1 and ICAM-1 proteins to improve the ability of the curcumin-loaded exosomes to penetrate the BBB [52].
Gene editing: Utilizing genetic engineering techniques could enable the specific expression of functional proteins on the cell membrane, which is difficult to achieve through surface chemical modifications. Since cancer cells exhibit low immunogenicity, cancer CMCDDSs must be combined with chemical immune adjuvants to enhance the immune response. Jiang et al. used gene editing to make cancer cell membranes express the CD80 protein and extracted engineered, cancer cell membranes to construct a cancer CMCDDS (Fig. 6B). The CD80 protein stimulates T-cell proliferation, and improves the effectiveness of tumor immunotherapy using CMCDDSs [53]. Rao et al. used gene editing to induce macrophage membrane surface overexpression of the SIRPα protein, which blocks the CD47-SIRPα pathway. The researchers extracted engineered, macrophage membranes to construct magnetic nanoparticles (gCM-MNs). The magnetic core nanoparticles promote M2 TAM repolarization and synergistically trigger potent macrophage immune responses [54].
In addition to participating in the regulation of the tumor immune microenvironment, gene-edited CMCDDSs also exhibit a strong ability to regulate the inflammatory microenvironment. Shi et al. used gene editing to make UVEC cell membranes express inflammatory factor receptors (TNFαR and IL-1R) and tumor necrosis factor-related apoptosis-inducing ligands (TRAILs). TRAIL ligands target the M1 macrophages in the inflammatory sites (Figs. 6C and D), while TNFαR and IL-1R can neutralize inflammatory factors. The same cell membrane gene editing strategies have also been applied in the treatment of myocardial infarction (MI) because they effectively inhibit the inflammatory response of MI [55].
The latest study used both external environmental-induced and gene editing-improved strategies, using genetic engineering to make cancer cell membrane surfaces express the SIRPα ligand, while using lipopolysaccharide (LSP) to induce M1 phenotype differentiation of macrophages. The researchers extracted the cell membranes of these two engineered cells and fused them with platelet membranes to create a multifunctional cell membrane material (hNVs), which effectively blocked the CD47-SIRPα pathway of macrophages, promoting M2-to-M1 repolarization within the tumor microenvironment. This hNVs CMCDDS specifically gathers at the surgical site, such that it can be used for the treatment of postoperative tumor recurrence [56].
3.2. Coating multi-functional core nanoparticlesCMCDDSs can effectively combine the advantages of cell membranes and chemical nanoparticles. In addition to functionalizing cell membranes, the core nanoparticles are not limited to materials such as polymers [20, 38, 57]. The range of core nanoparticles for CMCDDSs can be extended to nanogels, silica particles, gold nanoparticles, iron oxide nanoparticles, metal organic frameworks natural proteins, and nucleic acids. These core nanoparticles utilize their own functions, such as high drug loading, high adsorption capacity, photothermal conduction, and nuclear magnetic imaging, which combined with the biological functions of cell membranes to expand the application range of CMCDDSs in photodynamic therapy (PDT), PTT, MRI, natural proteins, and nucleic acid intracellular delivery. Each type of multi-functional core nanoparticles has its own advantages (Table 3) [58-81].
|
|
Table 3 Application of the multi-functional core nanoparticles in CMCDDS researches. |
Solution problems of drugs are a major issue affecting the industrial production. Nanocrystals, as solid, nanosized, drug particles, are simple to prepare, and covered with a layer of stabilizer that improves the solubility of insoluble drugs; nanocrystals are viewed as "pure particles of drug" with high drug loading capacity [82, 83]. Studies that combined nanocrystals with an erythrocyte membrane drug delivery system showed that the nanocrystals improved docetaxel solubility and safety, and the ligand-modified erythrocyte membranes improved docetaxel nanocrystal accumulation at the tumor sites [45].
3.3. Improving in vivo cell membrane smart shedding and drug release performanceEndogenous cell membranes can help CMCDDSs to avoid identification by the immune system, but whole cell membranes also limit drug release in target cells, and hinder uptake by target cells. The cell membrane material is thus a double-edged sword for the release of loaded drugs [84, 85]. Therefore, after endocytosis of the CMCDDS by the target cells, effective promotion of cell membrane smart shedding and drug release is an important factor affecting therapeutic efficacy. Previous studies have utilized different microenvironments at disease sites and external environmental stimuli to enable intelligent release of CMCDDSs.
3.3.1. pH-responsive CMCDDSsThe pH-sensitive response has been extensively researched in the field of intelligent drug delivery [86-88]. This response uses environmental pH changes at the disease site to regulate drug release. Zhang et al. designed a macrophage CMCDDS, that actively targets tumors. In the tumor microenvironment (pH 6.5), this system uses the proton sponge effect to cause an inflow of chloride ions and water from the tumor microenvironment, which finally leads to cell membrane delamination [86, 89]. When the core nanoparticles enter the intracellular microenvironment (pH 5.0), rapid release of the loaded drug is enabled (Figs. 7A and B). Liu et al. prepared pH-sensitive, liposomal POEz by fusing platelet membranes with pH-sensitive, lipid materials. POEz can respond to both the tumor microenvironment and lysosomal pH changes, which breaks the fusion membrane, the shedding of the platelet membrane, and causes the smart release of drugs that inhibits tumor cell growth [28]. CMCDDS studies on pH-responsive, intelligent release are often combined with PTT to improve treatment efficacy [90]. Song et al. developed an erythrocyte membrane camouflage nanogel (NRP+1) with tumor microenvironment responsive property. The NRP+1 can achieve high loading capability of chemotherapeutic drug (IL-2 and PTX). In pH 6.5, the erythrocyte membrane will be shed. These chemotherapeutic drugs were released to induce the expression of calreticulin on the surface of tumor cells, which can promote immunogenic cell death (ICD) [91].
|
Download:
|
| Fig. 7. (A) Illustration of the release mechanism of a pH-sensitive line bionic drug delivery system. (B) DLS, TEM imaging, and ζ potential changes of cell membrane bionic drug delivery system under different pH environments. Reproduced with permission [89]. Copyright 2018, American Chemical Society. (C) Fabrication process of biomimetic ROS-responsive nanocarriers co-loaded with Ce6 and TPZp. and Schematic illustration of NPs@i-RBMCe6+TPZp for tumortargeted PDT and hypoxia-activated chemotherapy upon light irradiation. (D) In vivo antitumor efficacy with laser irradiation (660 nm, 0.5 W/cm2, 10 min) in 4T1 tumor-bearing mice. Reproduced with permission [92]. Copyright 2019, Royal Society of Chemistry. | |
Studies have utilized tumor and inflammation sites that are high in reactive oxygen species (ROS) to design a variety of ROS-responsive CMCDDS. Liu et al. utilized ROS-sensitive nanocarriers loaded with the photosensitizer, Ce6, and the ROS-sensitive prodrug, tirapazamine (TPZp). The surface coating of functionally-modified RBC membranes enables the targeting of tumor cells. Upon light irradiation, Ce6, releases a large amount of ROS, which causes the destruction of ROS-sensitive nanocarriers and simultaneous TPZp activation and release (Figs. 7C and D) [92]. Gao et al. designed and synthesized a ROS-responsive material (Oxi-COS) that can self-assemble to form nanoparticles. Oxi-COS nanoparticles coated by macrophage membranes (MM-NPs) have the ability to target the site of inflammation [93]. Studies have shown that MM-NPs can release drugs rapidly in a high-ROS concentration environment. In the light of the high ROS concentration environment of ischemic stroke, researchers designed a ROS-sensitive erythrocyte membrane camouflage drug delivery system (SHp-RBC-NP) for stroke delivery of a neuroprotective agent (NR2B9C). After SHp-RBC-NP entered the brain site, it made full use of stroke homing peptide to achieve active targeting of ischemic injury, a high concentration of ROS leads to rapid release of NR2B9C [94].
3.3.3. External environmental stimulus responseExternal environmental stimuli can be used to achieve the smart release of CMCDDSs. This strategy has been mainly applied to sonodynamic therapy (SDT) and PDT. Feng et al. developed an MCF-7 cell membrane-coated drug delivery system (Figs. 8A and B). Under the action of external ultrasound, the MCF-7 cell membrane is shed [95]. Li et al. designed a cascade bioreactor (mCGP) to starve cancer cells. Glucose oxidase (GOx) and catalase are embedded in the surface of the cancer cell membrane, and coated with the porphyrin metal organic framework (Figs. 8C and D). Under light irradiation, intracellular hydrogen peroxide (H2O2) produces O2 and H2O, where O2 accelerates the decomposition of glucose and H2O promotes the proton sponge effect, leading to the breakup of the cell membrane material for smart shedding, and finally enhances the production of cytotoxic singlet oxygen (1O2) [96]. Hu et al. designed a M1-type macrophage membrane (M1m) camouflaged drug delivery system [(C/I)BP@B-A(D) & M1m]. After [(C/I)BP@B-A(D) & M1m] arrived the tumor site, the PEGylated bilirubin nanoparticles (BP) will be shed under exogenous laser irradiation, which releases the antitumor drug DOX, the photosensitizer Ce6, and the tumor immune reversal agent (indoleamine 2, 3-dioxygenase inhibitor). In this intelligent drug delivery system, the DOX and indoleamine 2, 3-dioxygenase inhibitor can synergistically promote tumor immune cell death. The Ce6 can generate large amounts of ROS under laser irradiation again, which further induces tumor cell death [97].
|
Download:
|
| Fig. 8. (A) Schematic mechanism of CCM-HMTNPs/HCQ for enhanced SDT on breast cancer via autophagy regulation strategy. (B) US irradiation time-dependent 1O2 generation of CCM-HMTNPs (20 μg/mL) determined using the singlet oxygen sensor green (SOSG) as an indicator and Release profiles of HCQ from CCM-HMTNPs/HCQ under US irradiation. Reproduced with permission [95]. Copyright 2019, American Chemical Society. (C) Schematic illustration of the cancer cell membrane camouflaged cascade bioreactor for cancer targeting starvation therapy and PDT. (D) The pH value changes of mCGP solution in the absence and presence of glucose and the O2 concentration changes of mCGP solution in the presence of H2O2 or glucose. Reproduced with permission. [96]. Copyright 2017, American Chemical Society. | |
In general, the stimuli that control the smart shedding and drug release of the CMCDDSs, it can be divided into endogenous or exogenous stimuli. Additionally, endogenous stimuli can be further partitioned into redox response, oxygen sensitivity, and bio-enzyme response [98]. Exogenous stimuli also include temperature-responsive, magnetic- responsive [99], can be combined with CMCDDS design to make CMCDDSs more intelligent, efficient, and accurate in the future.
4. Frontier medical applications of CMCDDSsCMBDDSs in anti-tumor research have been summarized by many articles [100-103]. In this review, we solely focus on medical frontier applications in anti-inflammatory treatments, anti-pathogenic microorganisms, and biological detoxification.
4.1. Anti-inflammatoryInflammation is usually associated with the immune system, which is involved in the occurrence of many inflammatory responses. In traditional nanomedicine design, targeted binding to inflammation through proteins on the cell membrane is often ignored [104]. By mimicking the interaction between the selectins or cellular adhesion molecules (CAMs) on inflammatory endothelial cells and leukocytes, CMCDDSs can realize inflammation-targeted drug delivery [105]. Immune cells, neutrophils, platelets, macrophages, stem cells, and other cell membranes are often used to prepare CMCDDSs, which have a wide range of applications in targeting, monitoring, and regulating inflammatory diseases. CMCDDSs have shown good therapeutic effects in inflammatory bowel disease [106], arterial tube stenosis [107], tumor inflammatory microenvironments [108], and sepsis [109]. Atherosclerosis, is the most common and deadly cardiovascular disease [34]. Wei et al. constructed a platelet membrane-camouflaged, PLGA, nanoparticle, biomimetic delivery, imaging system. The researchers utilized the ability of platelet membranes to specifically bind to inflammatory endothelial cells and subendothelial collagen matrices, which can realize the targeting of atherosclerotic sites, thus providing real-time medical imaging and facilitating clinical detection of the development of atherosclerosis [110].
4.2. Anti-pathogenic microorganismsIn recent years, due to the emergence of drug resistance, many bacterial infections have become incurable [111, 112]. Approximately 700, 000 people die from antibiotic resistance every year in the world. However, the development of new antibiotics is slow and cannot cope with the increasingly serious problem of resistance. Most pathogenic microorganisms interact with cells to produce toxicity [113, 114]. Pathogenic microorganisms enter the body and bind to normal cells (e.g., red blood cells [115]) to release toxin factors, which are also recognized and captured by immune cells (e.g., macrophages [116] and centrophages [117]). Therefore, CMCDDSs use cell membrane material surface-specific proteins to bind to microorganisms. Based on the above, CMCDDSs have provided many effective, therapeutic strategies against pathogenic microorganisms in recent years.
Anti-adhesion therapy interferes with the adhesion of bacteria to the host, which may slow down the development of drug resistance. Zhang et al. prepared an anti-adhesion cell membrane camouflaged platform by wrapping PLGA core nanoparticles with a Helicobacter pylori outer membrane, and the resulting bacterium-mimicking nanoparticles (OM-NPs) were produced (Figs. 9A and B). The experiments confirmed that OM-NPs have a high-binding ability with AGS cells and can compete with the source bacteria to bind the host [118]. RBC cell membrane-camouflaged drug delivery systems can the fungal production of hemolysis toxins through erythrocyte membrane surface proteins [21].

|
Download:
|
| Fig. 9. (A) Schematic representation of using OM-NPs to inhibit H. pylori adhesion on stomach lining. (B) Pretreatment of AGS cells with OM-NPs reduces subsequent fluorescence image and fluorescence quantification of H. pylori adhesion. Reproduced with permission [118]. Copyright 2019, Wiley Publishing Group. (C) Schematic representation of T-cell-membrane-coated nanoparticles (denoted as "TNPs") designed for attenuating HIV infectivity. (D) Inhibition of viral infection was tested with various TNP concentrations on each strain. Reproduced with permission [119]. Copyright 2018, Wiley Publishing Group. (E) Schematic mechanism of cellular nanosponges inhibiting SARS-CoV-2 infectivity. (F) Epithelial-NS, MΦ-NS and RBC-NS cellular nanosponges neutralize SARS-CoV-2 infectivity. Reproduced with permission [120]. Copyright 2020, American Chemical Society. | |
To improve the treatment and prevention of human immunodeficiency virus (HIV), it is very important to develop effective treatment strategies against broad-spectrum viral infections. Inspired by the recent development of cell membrane coating technology, Wei et al. developed a CD4+ T CMCDDS (TNPs). The CD4+ T cell membrane can specifically bind with HIV, which can transfer HIV from their intended host targets, thereby neutralizing HIV (Figs. 9C and D). This study has shown that TNPs can effectively neutralize HIV viral infections of monocytes and macrophages [119].
In 2020, COVID-19 has ravaged the world. So far, the global death toll has exceeded 2 million, making it a serious threat to the health of people around the world. Zhang et al. coated nanoparticles with the cell membrane of human lung epithelial cells or macrophages. These cell membrane materials carried the angiotensin-converting enzyme 2 (ACE2) and CD147 protein to combine with COVID-19 (Figs. 9E and F). Therefore, for the coronavirus, these cell membrane-camouflaged nanoparticles are not different from normal cells. The experiments showed that macrophage membranes reduce the infectivity of viruses by 88% and lung epithelial cell membrane viruses by 93% when using normal cell membrane-camouflaged nanoparticles at a concentration of 5 mg/mL [120].
4.3. Biological detoxificationIn addition to the targeting of pathogenic sites, CMCDDSs have a wide range of applications in biological detoxification. The principle is similar to that of anti-pathogenic microorganisms but it focuses more on the adsorption of toxin factors. After the concept of nano-sponge was proposed, erythrocyte membrane camouflaged drug delivery systems are the most extensively researched, due to erythrocyte membrane is capable of non-specific "uptake" of a variety of pore-causing toxins [121]. We believe that the main reason is that erythrocyte membrane camouflaged nanosponge is a kind of detoxification system that can adsorb broad spectrum of pore-forming toxins in nonspecific way. Organophosphorus, as the main component of pesticides, can lead to irreversible phosphorylation and the inactivation of acetylcholinesterase; the accumulation of acetylcholine in the body causes irreversible neuromuscular toxicity [122]. Acetylcholinesterase is expressed on the surface of RBC membranes. Pang et al. constructed an erythrocyte membrane-camouflaged drug delivery system, which reduces organophosphorus toxicity and combines with organophosphorus until they are smoothly metabolized in the liver [123]. Based on this, Chen et al. designed a cell-membrane-cloaked oil nanosponge, which consists of an olive oil and RBC membrane. The olive oil nonspecifically soaks up toxicants and the RBC membrane absorbs and neutralizes poisons through biological binding to the proteins [124]. Researchers discovered an erythrocyte membrane camouflage drug delivery system (RCB-NC), which can treat severe MRSA infections such as MRSA bacteremia and MRSA-induced sepsis [115]. In another example, Li et al. designed a platelet membrane camouflaged nano-robot (PL-motors). It inherits many of the properties of platelet membranes, which can adhesion and binding to toxins in whole blood, such as Shiga toxin and Staphylococcus aureus [125]. More importantly, the combination of cell membrane camouflaged technology and nanorobot technology provides a new idea for the future cell membrane camouflaged detoxification system.
5. Conclusions and perspectivesCMBDDS have gradually evolved into functional platforms gradually evolved into have CMBDDSs due to their unique transport kinetics, antigen spectrum, immunostimulatory properties, and targeting abilities for cells. Thus, CMCDDSs have unlimited possibilities in disease treatment. The CMBDDS has shown excellent treatment effectiveness for diseases that have been the focus of attention in the global medical community in recent years (e.g., reduction of pathogenic microbial resistance, HIV, and COVID-19), further increasing the confidence of researchers in its future development.
Personalized treatment has become an important development trend for future medical treatments. Increasing amounts of clinical data have found that there are individual differences in the disease status and the microenvironment of the disease, even if the patients have the same disease [121]. Consequently, it is necessary to develop customizable, personalized treatment methods. With the rapid development of individualized, immune cell therapy technology, this is made possible through genetic engineering modification of patients' immune cells, massive culture in vitro, and reinfusion into patients' bodies. Cellular immunotherapy technology is excellent for the personalized treatment of tumors and realizing "one-to-one", customizable, personalized treatment for patients. Based on development prospects in clinical personalized therapy, CMCDDSs are a drug delivery system derived from cells, where the majority of cellular biological functions are performed by cell membrane surface proteins [126]. Therefore, CMCDDSs also exhibit promising developments in clinical personalized treatment. Especially in the personalized treatment of tumors, the related studies have shown that the construction of a drug delivery system using the patients' own tumor cell can help to overcome the influence of the diversity of the tumor microenvironment of different individuals. Meanwhile, CMCDDSs can present complete tumor cell antigens to the immune system to realize the personalized treatment of patients' tumor sites. A study has found that the utilization of patient's head and neck squamous cell carcinoma membranes to construct a CMCDDS can overcome the influence of different, individual, tumor, microenvironment diversity, and can submit complete tumor cell antigens to the immune system to achieve personalized treatment for patient tumor sites. In addition, the drug delivery system is suitable for the treatment of unresectable tumors and metastatic tumors. Moreover, the therapeutic effect is obvious when combined with anticancer cisplatin [127].
Immune cell personalized therapy also has some challenges that affect its further development, such as the solid tumor immune microenvironment that suppresses the function of reinfusion immune cells, and the fact that solid tumors lack specific antigens. Therefore, personalized treatment of immune cells should be more inclined towards the organic combination of a variety of treatment methods to form a multi-functional compound treatment in the future. We believe that CMCDDSs, on their own as a personalized, therapeutic, can also be combined with immune cell therapies to constitute a novel cell therapy-cell membrane camouflaged drug delivery platform. This platform has certain development potential and will contribute to the innovation of tumor treatment methods and significantly improve the existing personalized tumor treatment methods.
In summary, functional CMCDDSs can increase the accumulation of drugs in pathogenic tissue, intelligently respond to the internal and external environment, release drugs, and optimize the comprehensive therapeutic effect. There is no doubt that the development of CMCDDSs will promote the progress of nanotherapy in the personalized treatment and diagnosis of diseases.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentsThis work was supported by the National Natural Science Foundation of China (No. 82073789) and Innovative Research Group at Higher Educational Institutions in Chongqing (No. CXQT20006).
| [1] |
L. Tang, Z. Wang, Q. Mu, et al., Adv. Mater. 32 (2020) 2002739. DOI:10.1002/adma.202002739 |
| [2] |
Y. Yang, Y. Yu, H. Chen, et al., ACS Nano 14 (2020) 13536-13547. DOI:10.1021/acsnano.0c05541 |
| [3] |
G. Chen, Y. Yang, Q. Xu, et al., Nano Lett. 20 (2020) 8141-8150. DOI:10.1021/acs.nanolett.0c03127 |
| [4] |
M. He, L. Yu, Y. Yang, et al., Chin. Chem. Lett. 31 (2020) 3178-3182. DOI:10.1016/j.cclet.2020.05.034 |
| [5] |
C.M.J. Hu, L. Zhang, S. Aryal, et al., Proc. Natl. Acad. Sci. U. S. A. 108 (2011) 10980-10985. DOI:10.1073/pnas.1106634108 |
| [6] |
R. Li, Y. He, Y. Zhu, et al., Nano Lett. 19 (2018) 124-134. |
| [7] |
L. Rao, L.L. Bu, B. Cai, et al., Adv. Mater. 28 (2016) 3460-3466. DOI:10.1002/adma.201506086 |
| [8] |
van der Meijden P.E.J., J.W.M. Heemskerk, Nat. Rev. Cardiol. 16 (2018) 166-179. |
| [9] |
A.M. Davy, H.F. Kildegaard, M.R. Andersen, Cell Syst. 4 (2017) 262-275. DOI:10.1016/j.cels.2017.02.010 |
| [10] |
A. Bauer, B. Kuster, Eur. J. Biochem. 270 (2003) 570-578. DOI:10.1046/j.1432-1033.2003.03428.x |
| [11] |
B.T. Luk, Jack Hu C., R.H. Fang, et al., Nanoscale 6 (2013) 2730-2737. DOI:10.1039/C3NR06371B |
| [12] |
Sancho-Albero M., Encabo-Berzosa M.d.M., Beltrán-Visiedo M., et al., Nanoscale 11 (2019) 18825-18836. DOI:10.1039/C9NR06183E |
| [13] |
M.J. Haney, Y. Zhao, Y.S. Jin, et al., J. Neuroimmune Pharmacol. 15 (2019) 487-500. |
| [14] |
Y. Li, J. Wu, J. Wang, et al., Acta Biomater. 101 (2020) 519-530. DOI:10.1016/j.actbio.2019.10.022 |
| [15] |
G. Liang, Y. Zhu, D.J. Ali, et al., J. Nanobiotechnol. 18 (2020) 10. DOI:10.1186/s12951-019-0563-2 |
| [16] |
B. Pang, Y. Zhu, J. Ni, et al., Theranostics 10 (2020) 2309-2326. DOI:10.7150/thno.39486 |
| [17] |
M.J. Haney, N.L. Klyachko, Y. Zhao, et al., J. Control. Release 207 (2015) 18-30. DOI:10.1016/j.jconrel.2015.03.033 |
| [18] |
S.C. Jang, O.Y. Kim, C.M. Yoon, et al., ACS Nano 7 (2013) 7698-7710. DOI:10.1021/nn402232g |
| [19] |
Y.T. Sato, K. Umezaki, S. Sawada, et al., Sci. Rep. 6 (2016) 21933. DOI:10.1038/srep21933 |
| [20] |
Z. Fan, H. Zhou, P.Y. Li, et al., J. Mater. Chem. B 2 (2014) 8231-8238. DOI:10.1039/C4TB00980K |
| [21] |
J. Xie, Q. Shen, K. Huang, et al., ACS Nano 13 (2019) 5268-5277. DOI:10.1021/acsnano.8b09681 |
| [22] |
C. Liu, J. Guo, F. Tian, et al., ACS Nano 11 (2017) 6968-6976. DOI:10.1021/acsnano.7b02277 |
| [23] |
L. Rao, B. Cai, L. Bu, et al., ACS Nano 11 (2017) 3496-3505. DOI:10.1021/acsnano.7b00133 |
| [24] |
Y. He, R. Li, H. Li, et al., ACS Nano 13 (2019) 4148-4159. DOI:10.1021/acsnano.8b08964 |
| [25] |
W. Liu, M. Ruan, Y. Wang, et al., Small 14 (2018) 1801754. DOI:10.1002/smll.201801754 |
| [26] |
A. Pitchaimani, T.D.T. Nguyen, S. Aryal, Biomaterials 160 (2018) 124-137. DOI:10.1016/j.biomaterials.2018.01.018 |
| [27] |
Z. Chen, P. Zhao, Z. Luo, et al., ACS Nano 10 (2016) 10049-10057. DOI:10.1021/acsnano.6b04695 |
| [28] |
G. Liu, X. Zhao, Y. Zhang, et al., Adv. Mater. 31 (2019) 1900795. |
| [29] |
K. Ding, C. Zheng, L. Sun, et al., Chin. Chem. Lett. 31 (2020) 1168-1172. DOI:10.1016/j.cclet.2019.10.040 |
| [30] |
W. Szlasa, I. Zendran, A. Zalesińska, et al., J. Bioenerg. Biomembr. 52 (2020) 321-342. DOI:10.1007/s10863-020-09846-4 |
| [31] |
X. Han, S. Shen, Q. Fan, et al., Sci. Adv. 5 (2019) eaaw6870. DOI:10.1126/sciadv.aaw6870 |
| [32] |
Q. Jiang, Y. Liu, R. Guo, et al., Biomaterials 192 (2019) 292-308. DOI:10.1016/j.biomaterials.2018.11.021 |
| [33] |
H. He, C. Guo, J. Wang, et al., Nano Lett. 18 (2018) 6164-6174. DOI:10.1021/acs.nanolett.8b01892 |
| [34] |
D. Dehaini, X. Wei, R.H. Fang, et al., Adv. Mater. 29 (2017) 1606209. DOI:10.1002/adma.201606209 |
| [35] |
G. Dou, R. Tian, X. Liu, et al., Sci. Adv. 6 (2020) eaba2987. DOI:10.1126/sciadv.aba2987 |
| [36] |
Q. Chen, G. Huang, W. Wu, et al., Adv. Mater. 32 (2020) 1908185. DOI:10.1002/adma.201908185 |
| [37] |
C. Qiu, H. Han, J. Sun, et al., Nat. Commun. 10 (2019) 2702. DOI:10.1038/s41467-019-10562-w |
| [38] |
F. Gao, L. Xu, B. Yang, et al., ACS Infect. Dis. 5 (2018) 218-227. |
| [39] |
L. Rao, Q. Meng, L. Bu, et al., ACS Appl. Mater. Interfaces 9 (2017) 2159-2168. DOI:10.1021/acsami.6b14450 |
| [40] |
H. Ding, Y. Lv, D. Ni, et al., Nanoscale 7 (2015) 9806-9815. DOI:10.1039/C5NR02470F |
| [41] |
Y. Guo, D. Wang, Q. Song, et al., ACS Nano 9 (2015) 6918-6933. DOI:10.1021/acsnano.5b01042 |
| [42] |
F. Gao, W. Li, J. Kan, et al., Adv. Biosyst. 4 (2020) 1900235. DOI:10.1002/adbi.201900235 |
| [43] |
F. Gao, W. Li, J. Deng, et al., ACS Appl. Mater. Interfaces 11 (2019) 18681-18690. DOI:10.1021/acsami.9b01725 |
| [44] |
Z. Chai, X. Hu, X. Wei, et al., J. Control. Release 264 (2017) 102-111. DOI:10.1016/j.jconrel.2017.08.027 |
| [45] |
Z. Chai, D. Ran, L. Lu, et al., ACS Nano 13 (2019) 5591-5601. DOI:10.1021/acsnano.9b00661 |
| [46] |
D. Zhu, W. Xie, Y. Xiao, et al., Nanotechnol. 29 (2018) 084002. DOI:10.1088/1361-6528/aa9ca1 |
| [47] |
H. Zhou, Z. Fan, P.K. Lemons, H. Cheng, Theranostics 6 (2016) 1012-1022. DOI:10.7150/thno.15095 |
| [48] |
X. Dong, L.L. Mu, X.L. Liu, et al., Adv. Funct. Mater. 30 (2020) 2000309. DOI:10.1002/adfm.202000309 |
| [49] |
C. Wang, Y. Wang, L. Zhang, et al., Adv. Mater. 30 (2018) 1804023. DOI:10.1002/adma.201804023 |
| [50] |
L. Pascucci, V. Coccè, A. Bonomi, et al., J. Control. Release 192 (2014) 262-270. DOI:10.1016/j.jconrel.2014.07.042 |
| [51] |
J. Wang, B.Z. Yeung, M. Cui, et al., J. Control. Release 268 (2017) 147-158. DOI:10.1016/j.jconrel.2017.10.020 |
| [52] |
H. Wang, H. Sui, Y. Zheng, et al., Nanoscale 11 (2019) 7481-7496. DOI:10.1039/C9NR01255A |
| [53] |
Y. Jiang, N. Krishnan, J. Zhou, et al., Adv. Mater. 32 (2020) 2001808. DOI:10.1002/adma.202001808 |
| [54] |
L. Rao, S.K. Zhao, C. Wen, et al., Adv. Mater. 32 (2020) 2004853. DOI:10.1002/adma.202004853 |
| [55] |
Y. Shi, F. Xie, P. Rao, et al., J. Control. Release 320 (2020) 304-313. DOI:10.1016/j.jconrel.2020.01.054 |
| [56] |
L. Rao, L. Wu, Z. Liu, et al., Nat. Commun. 11 (2020) 4909. DOI:10.1038/s41467-020-18626-y |
| [57] |
M. Gao, C. Liang, X. Song, et al., Adv. Mater. 29 (2017) 1701429. DOI:10.1002/adma.201701429 |
| [58] |
L. Li, J. Xu, G. Qi, et al., ACS Nano 8 (2014) 4975-4983. DOI:10.1021/nn501040h |
| [59] |
C. Gao, Z. Lin, Jurado-Sánchez B., et al., Small 12 (2016) 4056-4062. DOI:10.1002/smll.201600624 |
| [60] |
J. Su, H. Sun, Q. Meng, et al., Theranostics 7 (2017) 523-537. DOI:10.7150/thno.17259 |
| [61] |
L. Rao, Q. Meng, Q. Huang, et al., Adv. Healthcare Mater. 5 (2016) 1420-1427. DOI:10.1002/adhm.201600303 |
| [62] |
M. Xuan, J. Shao, L. Dai, et al., Adv. Healthcare Mater. 4 (2015) 1645-1652. DOI:10.1002/adhm.201500129 |
| [63] |
W. Gao, C.J. Hu, R.H. Fang, et al., Adv. Mater. 25 (2013) 3549-3553. DOI:10.1002/adma.201300638 |
| [64] |
J. Piao, L. Wang, F. Gao, et al., ACS Nano 8 (2014) 10414-10425. DOI:10.1021/nn503779d |
| [65] |
T. Jiang, B. Zhang, S. Shen, et al., ACS Appl. Mater. Interfaces 9 (2017) 31497-31508. DOI:10.1021/acsami.7b09458 |
| [66] |
L. Rao, L. Bu, J. Xu, et al., Small 11 (2015) 6225-6236. DOI:10.1002/smll.201502388 |
| [67] |
X. Ren, R. Zheng, X. Fang, et al., Biomaterials 92 (2016) 13-24. DOI:10.1016/j.biomaterials.2016.03.026 |
| [68] |
J. Guo, Y. Yu, W. Zhu, et al., Adv. Funct. Mater. 31 (2020) 2005935. |
| [69] |
J. Zhuang, Y. Duan, Q. Zhang, et al., Nano Lett. 20 (2020) 4051-4058. DOI:10.1021/acs.nanolett.0c01654 |
| [70] |
S. Li, B. Xie, H. Cheng, et al., Biomaterials 151 (2018) 1-12. DOI:10.1016/j.biomaterials.2017.10.021 |
| [71] |
S. Li, H. Cheng, W. Qiu, et al., Biomaterials 142 (2017) 149-161. DOI:10.1016/j.biomaterials.2017.07.026 |
| [72] |
D. Zhang, Z. Ye, L. Wei, et al., ACS Appl. Mater. Interfaces 11 (2019) 39594-39602. DOI:10.1021/acsami.9b14084 |
| [73] |
J. Zhuang, H. Gong, J. Zhou, et al., Sci. Adv. 6 (2020) eaaz6108. DOI:10.1126/sciadv.aaz6108 |
| [74] |
L. Chen, X. Zhao, Y. Liu, X. Yan, J. Mater. Chem. B 8 (2020) 8071-8083. DOI:10.1039/D0TB01272F |
| [75] |
Y. Xiao, W. Huang, D. Zhu, et al., RSC Adv. 10 (2020) 7194-7205. DOI:10.1039/C9RA09233A |
| [76] |
C. Gong, J. Tian, Z. Wang, et al., J. Nanobiotechnol. 17 (2019) 93. DOI:10.1186/s12951-019-0526-7 |
| [77] |
J. Yang, S. Wu, L. Hou, et al., Mol. Ther.-Nucleic Acids 21 (2020) 512-522. DOI:10.1016/j.omtn.2020.06.013 |
| [78] |
R. Chen, H. Huang, H. Liu, et al., Small 15 (2019) 1902686. DOI:10.1002/smll.201902686 |
| [79] |
L. Gao, L. Han, X. Ding, et al., Nanotechnol. 28 (2017) 335101. DOI:10.1088/1361-6528/aa7c43 |
| [80] |
X. Pang, X. Liu, Y. Cheng, et al., Adv. Mater. 31 (2019) 1902530. DOI:10.1002/adma.201902530 |
| [81] |
X. Liu, C. Liu, Z. Zheng, et al., Adv. Mater. 31 (2019) 1808294. DOI:10.1002/adma.201808294 |
| [82] |
L. Peltonen, J. Hirvonen, Int. J. Pharm. 537 (2018) 73-83. DOI:10.1016/j.ijpharm.2017.12.005 |
| [83] |
P. Maudens, C.A. Seemayer, F. Pfefferlé, et al., J. Control. Release 276 (2018) 102-112. DOI:10.1016/j.jconrel.2018.03.007 |
| [84] |
S. Aryal, C.J. Hu, R.H. Fang, et al., Nanomedicine 8 (2013) 1271-1280. DOI:10.2217/nnm.12.153 |
| [85] |
R.H. Fang, C.J. Hu, B.T. Luk, et al., Nano Lett. 14 (2014) 2181-2188. DOI:10.1021/nl500618u |
| [86] |
W. Chen, K. Zeng, H. Liu, et al., Adv. Funct. Mater. 27 (2017) 1605795. DOI:10.1002/adfm.201605795 |
| [87] |
P. Zheng, Y. Liu, J. Chen, et al., Chin. Chem. Lett. 31 (2020) 1178-1182. DOI:10.1016/j.cclet.2019.12.001 |
| [88] |
X. Li, Y. Zhang, Z. Ma, et al., Chin. Chem. Lett. 30 (2019) 489-493. DOI:10.1016/j.cclet.2018.03.019 |
| [89] |
Y. Zhang, K. Cai, C. Li, et al., Nano Lett. 18 (2018) 1908-1915. DOI:10.1021/acs.nanolett.7b05263 |
| [90] |
S. Ye, F. Wang, Z. Fan, et al., ACS Appl. Mater. Interfaces 11 (2019) 15262-15275. DOI:10.1021/acsami.9b00897 |
| [91] |
Q. Song, Y. Yin, L. Shang, et al., Nano Lett. 17 (2017) 6366-6375. DOI:10.1021/acs.nanolett.7b03186 |
| [92] |
H. Liu, W. Jiang, Q. Wang, et al., Biomater. Sci. 7 (2019) 3706-3716. DOI:10.1039/C9BM00634F |
| [93] |
C. Gao, Q. Huang, C. Liu, et al., Nat. Commun. 11 (2020) 2622. DOI:10.1038/s41467-020-16439-7 |
| [94] |
W. Lv, J. Xu, X. Wang, et al., ACS Nano 12 (2018) 5417-5426. DOI:10.1021/acsnano.8b00477 |
| [95] |
Q. Feng, X. Yang, Y. Hao, et al., ACS Appl. Mater. Interfaces 11 (2019) 32729-32738. DOI:10.1021/acsami.9b10948 |
| [96] |
S. Li, H. Cheng, B. Xie, et al., ACS Nano 11 (2017) 7006-7018. DOI:10.1021/acsnano.7b02533 |
| [97] |
C. Hu, T. Lei, Y. Wang, et al., Biomaterials 255 (2020) 120159. DOI:10.1016/j.biomaterials.2020.120159 |
| [98] |
S. Mura, J. Nicolas, P. Couvreur, Nat. Mater. 12 (2013) 991-1003. DOI:10.1038/nmat3776 |
| [99] |
A. Raza, T. Rasheed, F. Nabeel, et al., Molecules 24 (2019) 1117. DOI:10.3390/molecules24061117 |
| [100] |
R. Li, Y. He, S. Zhang, et al., Acta Pharm. Sin. B 8 (2018) 14-22. DOI:10.1016/j.apsb.2017.11.009 |
| [101] |
Z. He, Y. Zhang, N. Feng, Mater. Sci. Eng. C 106 (2020) 110298. DOI:10.1016/j.msec.2019.110298 |
| [102] |
Y. Zhou, W. Zhou, X. Chen, et al., Acta Pharm. Sin. B 10 (2020) 1563-1575. DOI:10.1016/j.apsb.2019.11.013 |
| [103] |
X. Zhen, P. Cheng, K. Pu, Small 15 (2019) 1804105. DOI:10.1002/smll.201804105 |
| [104] |
K. Jin, Z. Luo, B. Zhang, Z. Pang, Acta Pharm. Sin. B 8 (2018) 23-33. DOI:10.1016/j.apsb.2017.12.002 |
| [105] |
H. Wang, Y. Liu, R. He, et al., Biomater. Sci. 8 (2020) 552-568. DOI:10.1039/C9BM01392J |
| [106] |
C. Corbo, W.E. Cromer, R. Molinaro, et al., Nanoscale 9 (2017) 14581-14591. DOI:10.1039/C7NR04734G |
| [107] |
C.J. Hu, R.H. Fang, K. Wang, et al., Nature 526 (2015) 118-121. DOI:10.1038/nature15373 |
| [108] |
Q. Hu, W. Sun, C. Qian, et al., Adv. Mater. 29 (2017) 1605803. DOI:10.1002/adma.201605803 |
| [109] |
R. Molinaro, A. Pastò, C. Corbo, et al., Nanoscale 11 (2019) 13576-13586. DOI:10.1039/C9NR04253A |
| [110] |
X. Wei, M. Ying, D. Dehaini, et al., ACS Nano 12 (2017) 109-116. |
| [111] |
L.J.V. Piddock, Nat. Rev. Microbiol. 15 (2017) 639-640. DOI:10.1038/nrmicro.2017.121 |
| [112] |
H. Chang, T. Cohen, Y.H. Grad, etal., Microbiol. Mol. Biol.Rev. 79 (2015) 101-116. DOI:10.1128/MMBR.00039-14 |
| [113] |
R.H. Fang, Y. Jiang, J.C. Fang, L. Zhang, Biomaterials 128 (2017) 69-83. DOI:10.1016/j.biomaterials.2017.02.041 |
| [114] |
R.H. Fang, B.T. Luk, C.J. Hu, L. Zhang, Adv. Drug Deliv. Rev. 90 (2015) 69-80. DOI:10.1016/j.addr.2015.04.001 |
| [115] |
Y. Chen, Y. Zhang, M. Chen, et al., Small 15 (2019) 1804994. DOI:10.1002/smll.201804994 |
| [116] |
X. Wei, D. Ran, A. Campeau, et al., Nano Lett. 19 (2019) 4760-4769. DOI:10.1021/acs.nanolett.9b01844 |
| [117] |
K. Wang, Y. Lei, D. Xia, et al., Colloids Surf. B 188 (2020) 110755. DOI:10.1016/j.colsurfb.2019.110755 |
| [118] |
Y. Zhang, Y. Chen, C. Lo, et al., Int. Ed. 58 (2019) 11404-11408. DOI:10.1002/anie.201906280 |
| [119] |
X. Wei, G. Zhang, D. Ran, et al., Adv. Mater. 30 (2018) 1802233. DOI:10.1002/adma.201802233 |
| [120] |
Q. Zhang, A. Honko, J. Zhou, et al., Nano Lett. 20 (2020) 5570-5574. DOI:10.1021/acs.nanolett.0c02278 |
| [121] |
J. Zhou, A.V. Kroll, M. Holay, et al., Adv. Mater. 32 (2019) 1901255. |
| [122] |
P. Masson, F. Nachon, J. Neurochem. 142 (2017) 26-40. DOI:10.1111/jnc.14026 |
| [123] |
Z. Pang, C.J. Hu, R.H. Fang, et al., ACS Nano 9 (2015) 6450-6458. DOI:10.1021/acsnano.5b02132 |
| [124] |
Y. Chen, Y. Zhang, J. Zhuang, et al., ACS Nano 13 (2019) 7209-7215. DOI:10.1021/acsnano.9b02773 |
| [125] |
J. Li, P. Angsantikul, W. Liu, et al., Adv. Mater. 30 (2018) 1704800. DOI:10.1002/adma.201704800 |
| [126] |
X. Gong, J. Li, T. Tan, et al., Adv. Funct. Mater. 29 (2019) 1903441. DOI:10.1002/adfm.201903441 |
| [127] |
L. Rao, G.T. Yu, Q.F. Meng, et al., Adv. Funct. Mater. 29 (2019) 1905671. DOI:10.1002/adfm.201905671 |
 2021, Vol. 32
2021, Vol. 32