b Wuhan No. 1 Huiquan Middle School, Wuhan 430022, China
Nucleic acids are associated with the accelerated aggregation of proteins including amyloid- β, prion protein (PrP), gelsolin, α-synuclein, β2-microglobulin, transthyretin and peptide hormones, which are implicated in a variety of diseases and biochemical processes [1, 2]. They are capable of binding to monomers and oligomers of proteins, likely via nonspecific interaction patterns, leading to the formation of proteinaceous inclusions [3]. These interactions with soluble protein oligomers facilitate rapid conversion into insoluble amyloid, and thereby diminish the cytotoxicity of the protein oligomers [2]. However, nucleic acid-containing amyloid fibrils induce type I interferon production and stimulate systemic autoimmunity [4]. The formation of PrP–nucleic acid complexes seems to accelerate the conversion of cellular forms of PrP into the disease-causing isoform [5]. Nonspecific interactions between PrP and two single-stranded DNA (ssDNA) oligomers also lead to different cytotoxic protein aggregates [6]. Although these reports show the inconsistent roles in regulation of aggregate cytotoxicity, the involvement of nucleic acids in the rapid formation of disease protein aggregates or inclusions might be a promising approach to regulate both formation and morphology of the protein aggregates, and thereby control cytotoxicity of the aggregates.
Nucleic acids are also necessary for formation of biological aggregates or condensates involved in intracellular phase transitions and neurodegeneration [7, 8]. One of the unique structures enriched in transcripts recruited to phase-separated condensates is G4-quadruplexes. Nucleic acid fragments with G4 elements have been shown to play a role in the formation of phase-separated aggregates [9]. Nucleic acid G4-quadruplexes have been identified for the hexanucleotide repeats [10].
Cu/Zn superoxide dismutase (SOD1) is an essential antioxidant enzyme widely distributed in cells, which catalyzes the rapid dismutation of O2·− into H2O2 and O2, and maintains the intracellular reactive oxygen species (ROS) homeostasis [11, 12]. Therefore, SOD1 has been reported to play an important roles in redox signaling [13, 14], nutrient sensing [15] and respiration [16] via controlling cellular ROS levels. Recently, it was reported that SOD1 can also bind to DNA regulating gene expression [17]. However, it is still unclear whether nucleic acids are involved in the formation of proteinaceous inclusions or deposits rich in SOD1 in cultured cells and animal model experiments, as well as in samples from amyotrophic lateral sclerosis patients so far. We previously found that wild-type SOD1 had an apparent affinity for DNA (Kd) of up to 0.5 mol/L at pH 3.6 and ~10 mol/L at pH 7.4 [18], and the conversion of SOD1 into aggregates was accelerated by DNA binding under acidic conditions, indicating a template effect of DNA in SOD1 aggregation [18-21]. In addition, we reported that the profiles of SOD1 aggregates could be regulated to a certain extent by different forms of double-stranded DNA with indefinite lengths [18-21]. Moreover, our results imply that sequences and secondary structures of DNAs determine both formation and morphology of SOD1 aggregates under the conditions tested [3]. Herein, we examined the regulatory roles in both morphology and cytotoxicity of SOD1 aggregates by four ssDNA oligomers (dN)12 (N=A, T, G, C) and their G-quadruplets under acidic conditions.
Indeed, biological systems generally resided in near neutral environment, and the aggregation of SOD1 under acidic condition but not neutral condition may affect their biological applications to some extent. However, there are still many biological activities reported to be involved in acidic environments in cells or organs, and the SOD1 aggregates might be applied. For instance, as reported in reference, lysosomes are involved in protein degradation at a unique physiological acidic pH range (pH 3.8–5.0) [22], and in the neural stem cell (NSC) pool, quiescent NSCs from young mice contained large lysosomes which stored many protein aggregates [23], SOD1 aggregates, on the one hand, could provide a mode for protein aggregation in biological research and for SOD1, and on the other hand, could potentially be used as a RNA and gene carrier. The stable and nontoxic SOD1–nucleic acid aggregates formed at pH 4.0, would gradually dissociate and release nucleic acid in the internal neutral environment.
In order to evaluate affinity of SOD1 for ssDNA oligomers under acidic conditions, the binding equilibrium constants were measured by isothermal titration calorimetry. The titration of ssDNA oligomers with SOD1 was carried out at pH 4.0, and injections occurred at 60-second intervals to allow equilibrium and avoid the interference from SOD1 aggregation in the binding. SOD1 aggregation was expected to be negligible because binding is much faster than aggregation [18-20]. Fig. S1 (Supporting information) indicated that a strong binding affinity (Kd) of ssDNA oligomers with SOD1 was existed [19].
These results prompted us to examine the changes in hydrophobic clusters of SOD1 caused by the binding of ssDNA oligomers. To this end, 8-anilino-1-napthalene-sulfonic acid (ANS) binding experiments were performed for SOD1 samples with varied concentrations of ssDNA oligomers. The fluorescence of ANS was identical to the fluorescence of (dN)12, whereas incubation with SOD1 for 2 h without ssDNA oligomers led to about 33% enhancement with a 20 nm blue shift of the strongest ANS fluorescence (Fig. S2 in Supporting information). These results indicated that SOD1 proteins exposure to the acidic environment for a long period could also lead to an increase in the hydrophobic surfaces of SOD1 and ANS could bond to SOD1. Unexpectedly, in the presence of both ssDNA oligomers and SOD1, the (dA)12 and (dT)12 had a 133% enhancement and a 40 nm blue shift of the strongest ANS emission relative to that of ANS in the buffer, while (dC)12 and (dG)12, led to more than two-fold enhancement and 40 nm blue shift of the strongest ANS emission (Fig. S2). These results demonstrated that the binding of ssDNA oligomers could significantly increase the exposure of hydrophobic clusters in SOD1. Binding of (dC)12 and (dG)12 resulted in much more pronounced conformational changes of SOD1 than that of (dA)12 and (dT)12.
Because increased hydrophobicity is a prerequisite for protein aggregation, transmission electron microscopy (TEM) were performed to exam whether SOD1 aggregates could formed upon incubation with ssDNA oligomers. SOD1 was incubated with varied concentrations of ssDNA oligomers at pH 4.0 for 2 h, 3 days and 7 days, respectively. In consistence with the results from the ANS binding experiments, the observed SOD1 aggregates could be classified into two types according to their profiles: the aggregates formed with (dA)12 or (dT)12 after incubation for 2 h were amorphous species with variable sizes, whereas those formed with (dC)12 or (dG)12 were fibril-like species or fibrillar with ~10 nm diameters and variable lengths (Fig. 1). And the profiles of SOD1 aggregates remained unchanged with incubation time.

|
Download:
|
| Fig. 1. TEM imaging for wild-type SOD1 (wt SOD1) aggregates formed with (dN)12 (N=A, T, C, G). Scale bar; 200 nm. (A–D) SOD1 (wt SOD1) aggregates formed with (dA)12 (A), (dT)12 (B), (dC)12 (C) and (dG)12 (D). (E) Proposed aggregation mechanism of SOD1 around (dG)12 at pH 4.0. The hydrophobic interactions occurred in aggregation are indicated by "≡". | |
To further support the morphological presumption, the fluorescence spectra of thioflavin T (ThT) interacting with SOD1 aggregates were measured due to its ability of binding fibrillar protein aggregates to enhance fluorescence. As expected, the fluorescence intensity of ThT raised significantly after incubated with SOD1 species in presence of (dG)12. While the fluorescence of ThT remained almost unchanged, when treated with SOD1 species along with (dA)12, (dC)12 or (dT)12 for instead. However, the control experiments indicated that (dG)12 without SOD1 still induced a similar fluorescence enhancement of ThT.
As ThT was also a typical and efficient fluorescent sensor for G-quadruplex [24], we therefore proposed that G-quadruplex would generated from (dG)12 under the tested conditions (pH 4.0) and induced the fluorescence enhancement of TnT (Fig. 2). On the contrary, (dA)12, (dC)12 and (dT)12 which were not able to form G-quadruplex showed little or even no fluorescent response at the same situation (Fig. 2A). While ThT may acted as G-quadruplex probe much more than fibrillar protein aggregates probe and therefore display minor response towards fibrillar SOD1 aggregates.

|
Download:
|
| Fig. 2. (A) Fluorescence spectra and time courses of the strongest fluorescence of ThT (excitation at 440 nm) in pH 4.0 buffer, 4 μmol/L SOD1- or 8 μmol/L (dN)12-containing buffer (pH 4.0), and solutions containing 4 μmol/L SOD1 and 8 μmol/L (dN)12 (pH 4.0), respectively. (B) Fluorescence spectra of the strongest fluorescence of TO (excitation at 501 nm) in pH 4.0 buffer, 4 μmol/L SOD1- or 8 μmol/L (dN)12-containing buffer (pH 4.0), and solutions containing 4 μmol/L SOD1 and 8 μmol/L (dN)12 (pH 4.0), respectively. | |
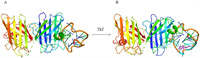
We also checked the docking model of SOD1 and (dG)12 complex with ThT by HADDOCK [25] and estimated its' affinity and interaction scores. The optimized combination mode of (dG)12 and SOD1 was shown in Fig. 3A with the lowest HADDOCK score (−210.3±3.7). When the molecule of ThT approached the G-quadruplex, it could bond to the G-quadruplex at the site of groove and the S atom of ThT would interact with the G6 from the G-quadruplx to form a strong hydrogen bond. And the fluorescence changes of ThT was mainly associated with angles between the two conjugated aomatic rings of ThT [26], the involvement of SOD1 may only affect the rotaion of benzothiazole and dimethylamino benzene of ThT (Fig. 3B), which lead to the just little change of fluorescence intensity of ThT. It was in consistent with the results in Fig. 2A.
|
Download:
|
| Fig. 3. (A) The docking model of SOD1 with (dG)12. (B) The docking model of SOD1 with (dG)12 and ThT. | |
We also carried out experiments by using thiazole orange (TO), another fluorescent probe widely used for detecting not only smart G-quadruplex binder dye but also protein aggregates, for instead [27] to further certify our speculation. We found that (dG)12 induced a rapid and enormous fluorescence enhancement of TO independently compared to other (dN)12, the involvement of SOD1 even diminished the fluoresence. We thought the formation of G-quadruplex increased the fluorescence intensity of TO from 3.74 to 537.35, and the formation of SOD1 fibrillar aggregates triggered another fluorescence enhancement from 2.75 to 945.82 (Fig. 2B). These results together with the observations of TEM indicated that the fibrillar SOD1 aggregates were formed when incubation with (dG)12, and G-quadruplex might play an important role in the fibrotic aggregation of SOD1.
For further confirming the formation of a DNA G-quadruplex in the system composed of SOD1 and (dG)12, we examined the structures of ssDNA oligomers using circular dichroism (CD) [28]. The conformational switch from the cytosine (C)-rich ssDNA to an i-motif structure occurs at acidic pH with two distinct peaks, at ~285 nm and ~260 nm, known as characteristic CD bands of i-motif DNAs [29]. A comparison of the CD spectra of (dC)12 measured, respectively, in buffer, SOD1 solution and water showed that the (dC)12 was converted into an i-motif structure both in buffer and in SOD1 solution as indicated by the strong peaks at 266 nm and 288 nm (Fig. 4). On the other hand, G-rich ssDNAs stack upon each other, leading to an intermolecular or intramolecular G-quadruplex structure [30] and it's all strands being parallel structure had a CD spectrum contains a maximum peak around 265 nm and a minimum peak around 240 nm [31]. CD peaks at both 263 nm and 240 nm were observed for the (dG)12 both in the buffer and in the SOD1 solution (Fig. 4), revealing that the (dG)12 in these states had a quadruplex structure. The structures of (dA)12 and (dT)12 in the SOD1 solutions were also examined by CD. Following the incubation either in buffer or in SOD1 solution at 37 ℃, the results of CD spectra were similar to each other (Fig. 4), indicated that (dA)12 and (dT)12 both in the buffer and in the amorphous aggregates remain a single-stranded flexible form [32]. Based on these results, we concluded that these four kinds of ssDNA molecules existed in the SOD1 aggregates had two conformational states, as indicated by their CD (Fig. S5 in Supporting information), which corresponded to two morphologies of SOD1 aggregates.

|
Download:
|
| Fig. 4. (A) CD spectra of (dA)12 in water, 50 mmol/L HAc-NaAc buffer (pH 4.0), and SOD1 aggregates (pH 4.0). (B) CD spectra of (dT)12 in water, 50 mmol/L HAc-NaAc buffer (pH 4.0) and SOD1 aggregates (pH 4.0). (C) CD spectra of (dC)12 in water, 50 mmol/L HAc-NaAc buffer (pH 4.0) and SOD1 aggregates (pH 4.0). (D) CD spectra of (dG)12 in water, 50 mmol/L HAc-NaAc buffer (pH 4.0) and SOD1 aggregates (pH 4.0). | |
Significant lags in the aggregation with ssDNA oligomers were not found in the kinetic traces of ThT fluorescence in addition to the dead time of 15 s prior to measurement (inset in Fig. 2). To confirm this accelerating role of ssDNA oligomers, time courses of incubation with (dN)12 under the tested conditions were also monitored with static light scattering (SLS) observations. A lag of several seconds appeared in the case of (dA)12 or (dT)12 and (dC)12 (Fig. S3 in Supporting information) while there's almost no lag for (dG)12. The lag time of several hours, days and even weeks has been reported for the aggregation of apo and mutant forms of SOD1 in some cases that required protein denaturing agents [33-35]. Thus, the ssDNA oligomers significantly accelerated SOD1 aggregation, and the presence of (dG)12 resulted in the rapid SOD1 aggregation with unobservable lags. Moreover, the acceleration is enhanced with increasing ratios of SOD1 to (dN)12 (Fig. S4 in Supporting information). On the other hand, the time traces monitored with both ThT binding and SLS indicated that the aggregation of SOD1 with ssDNA oligomers was dominated by two kinetic phases. In the first phase, occurring on a time scale of a decade of seconds, the ThT fluorescence dramatically increased. This fast phase was followed by a slow phase in which the ThT fluorescence and SLS remained unchanged.
The polymorphism of SOD1 aggregates formed with four kinds of ssDNA molecules could reasonably be explained by their conformations in the SOD1 aggregates and in the buffer. The binding of ssDNA oligomers caused a conformational alteration of SOD1, leading to the exposure of more hydrophobic clusters in the protein. The intermolecular hydrophobic interactions of SOD1 promoted the protein aggregation along the single chain ssDNAs. The granular SOD1 aggregates were generated from the association with (dA)12 or poly(dT)12 in the randomly coiled conformational state because of their flexiblity. The formation of large and amorphous SOD1 species could be attributed to the interactions among the nanoparticles, as delineated in our previous studies [18, 19].
Furthermore, our results indicated that the assembly with (dG)12 resulted in the formation of fibrillar SOD1 aggregates (Fig. 1), whereas the assembly around the (dC)12 i-motifs resulted in the formation of short and loose species, as observed under TEM (Fig. 1). In fact, the protein fibrils formed with (dG)12 were much longer and straighter than those with (dC)12 (Fig. 1), which could be ascribed to the fact that (dG)12 could readily produce a long and linear G-quadruplex, but (dC)12 could not. In addition, the intermolecular hydrophobic interactions of SOD1 at the ends of the fibril-like (also fibrillar) species may have contributed to the formation of longer fibrils.
The semi-quantitative analysis based on the previously reported structural data might provide further insight into the mechanism of aggregation of SOD1 with (dG)12. The X-ray structural determination revealed that three mutant SOD1 proteins produced the amyloid-like filaments with diameters of 9.5 nm. The inside of each filament was a square water-filled nanotube with a diagonal length of 3.0 nm (Fig. 1) [36]. TEM showed that the fibrillar SOD1 aggregates had diameters of ~10 nm according to the scale bar (Fig. 1), whereas the (dG)12 had diameters of ~3.0 nm [37]. These data demonstrated that (1) the fibrillar SOD1 aggregates formed with or without (dG)12 had similar diameters, (2) the G-quadruplex could fill the nanotube of the filaments based on their well-matched sizes, and (3) the hydrophobic interactions to drive aggregation were the same for aggregation with or without (dG)12. The G-quadruplex acted as templates that concentrated SOD1 molecules around themselves by electrostatic attraction and promoted the intermolecular hydrophobic contacts of SOD1 (Fig. 1).
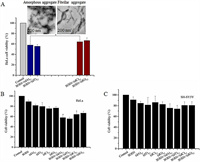
Finally, cytotoxicity of the SOD1 aggregates formed with ssDNA oligomers at the given concentrations was evaluated by MTT relative cell viability assay [38]. After SH-SY5Y and HeLa cells were exposed to the aggregates for 24 h, the MTT data showed that the relative viability of cells exposed to the SOD1 aggregates formed with (dC)12 or (dG)12 was close to that of cells exposed to (dN)12 (80% for SH-SY5Y, 68% for HeLa), but lower than that of cells exposed to the freshly prepared SOD1 (90%) (Fig. 5). In contrast, the SOD1 aggregates with (dA)12 or (dT)12 reduced viability (72% for SH-SY5Y and 58% for HeLa).
|
Download:
|
| Fig. 5. (A) Structures of SOD1 aggregates influenced relative viability which evaluated by MTT on HeLa cells. Data are expressed as mean±SD (n=5). *P < 0.05. (B, C) Relative viability evaluated by MTT on HeLa and SH-SY5Y cells. Data are expressed as mean ± SD (n=5).*P < 0.05. | |
These data indicated that (1) the growth of the neuron-like cells is not significantly affected by the ssDNA oligomer-containing SOD1 aggregates, (2) the cells exposed to the (dC)12- or (dG)12-containing SOD1 aggregates exhibited relatively high viability, while cells exposed to the (dA)12- or (dT)12-containing SOD1 aggregates exhibited poor viability, and (3) HeLa cells are more vulnerable to the ssDNA-containg SOD1 aggregates than SH-SY5Y cells. Based on these results, we concluded that the amorphous SOD1 species have higher cytotoxicity than the fibrillar and fibril-like SOD1 species. This raises a possibility that the cytotoxicity of protein aggregates could be regulated by ssDNAs if ssDNAs can act as an accelerating template in protein aggregation. On the other hand, the relative viability was reported to be less than 10% after exposure to the aggregates of apoSOD1 (on PC-12 cells) and mutant SOD1 proteins (on HEK293 cells) [39, 40]. Therefore, the cytotoxicity of SOD1 aggregates with ssDNA oligomers is much less than that of apoSOD1 and mutant SOD1 aggregates with no nucleic acid. In addition, we found that the relative viability of HeLa cells was lower than that of SH-SY5Y cells following treatment with the ssDNA-containing SOD1 aggregates (Fig. 5), indicating that the SOD1 aggregates were more toxic to HeLa cells than to motor neuron-like cells.
In conclusion, the models of SOD1 binding with ssDNAs oligomers were conducted, producing different kinds of aggregates in the case of flexible single or rigid quadruple ssDNAs. These results provide not only additional evidence to support the template role of nucleic acids in the aggregation of SOD1 proteins, which was proposed in our previous studies [19, 21], but also help us understand roles of nucleic acid G-quadruplexes in the formation of condensates and membraneless organelles involved in intracellular phase transitions. The profile-dependent cytotoxicty of SOD1 aggregates suggests a possibility that the cytotoxicity of protein aggregates could be regulated by protein-bound DNAs.
Declaration of competing interestThe authors report no declarations of interest.
AcknowledgmentsWe thank Professor Hongzhe Sun at Hong Kong University for his helpful comments and technical support and my colleague Professor Soumyajit Roy for reversion of this manuscript. This work was financially supported by the National Natural Science Foundation of China (Nos. 21771073, 22077046, 21001047, 21072074) and the Fundamental Research Funds for the Central Universities (No. CCNU19TS052).
Appendix A. Supplementary dataSupplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.cclet.2021.01.045.
| [1] |
M. Calamai, J.R. Kumita, J. Mifsud, et al.
, Biochem. 45 (2006) 12806-12815. DOI:10.1021/bi0610653 |
| [2] |
J.D. Domizio, R. Zhang, L.J. Stagg, et al., J. Biol. Chem. 287 (2012) 736-747. DOI:10.1074/jbc.M111.238477 |
| [3] |
C.L. Liu, Y. Zhang, Adv. Protein Chem. Struct. Biol. 84 (2011) 1-40. DOI:10.1145/2037715.2037759 |
| [4] |
J.D. Domizio, S. Dorta-Estremera, M. Gagea, et al., Proc. Natl. Acad. Sci. U. S. A. 109 (2012) 14550-14555. DOI:10.1073/pnas.1206923109 |
| [5] |
C.N. Wong, L.W. Xiong, M. Horiuchi, et al., EMBO J. 20 (2001) 377-386. DOI:10.1093/emboj/20.3.377 |
| [6] |
B. Macedo, T.A. Millen, C.A.C.A. Braga, et al., Biochem. 51 (2012) 5402-5413. DOI:10.1021/bi300440e |
| [7] |
S.F. Banani, A.M. Rice, W.B. Peeples, et al., Cell 166 (2016) 651-663. DOI:10.1016/j.cell.2016.06.010 |
| [8] |
A. Khong, T. Matheny, S. Jain, et al., Cell 68 (2017) 808-820. DOI:10.1016/j.molcel.2017.10.015 |
| [9] |
P. Ivanov, E. O'Day, M.M. Emara, et al., Proc. Natl. Acad. Sci. U. S. A. 111 (2014) 18201-18206. DOI:10.1073/pnas.1407361111 |
| [10] |
Y. Zhang, C. Roland, C. Sagui, Neurosci. 9 (2018) 1104-1117. DOI:10.1021/acschemneuro.7b00476 |
| [11] |
X. Li, J.Y. Shi, S. Qiu, et al., Prog. Chem. 30 (2018) 1475-1486. |
| [12] |
X. Li, Y. Chen, J. Zhao, et al., Oxid. Med. Cell. Longev. 2019 (2019) 9706792. |
| [13] |
B.J. Carter, P. Anklesaria, S. Choi, et al., Antioxid. Redox Sign. 11 (2009) 1569-1586. DOI:10.1089/ars.2008.2414 |
| [14] |
A.E. Frakes, L. Ferraiuolo, A.M. Haidet-Phillips, et al., Neuron 81 (2014) 1009-1023. DOI:10.1016/j.neuron.2014.01.013 |
| [15] |
X. Li, S. Qiu, J. Shi, et al., Nucleic Acids Res. 47 (2019) 5074-5085. DOI:10.1093/nar/gkz256 |
| [16] |
C.K. Tsang, M. Chen, X. Cheng, et al., Mol. Cell 70 (2018) 502-515. DOI:10.1016/j.molcel.2018.03.029 |
| [17] |
A.R. Reddi, V.C. Culotta, Cell 152 (2013) 224-235. DOI:10.1016/j.cell.2012.11.046 |
| [18] |
W. Jiang, Y.C. Han, R.Y. Zhou, et al., Biochem. 46 (2007) 5911-5923. DOI:10.1021/bi062234m |
| [19] |
W. Jiang, B. Zhang, J. Yin, et al., Biopolymers 89 (2008) 1154-1169. DOI:10.1002/bip.21067 |
| [20] |
J. Yin, S. Hu, W. Jiang, et al., PLoS One 5 (2010) e12328. DOI:10.1371/journal.pone.0012328 |
| [21] |
J. Yin, R.Z. Chen, C.L. Liu, Front. Biosci. 14 (2009) 5084-5106. DOI:10.2741/3588 |
| [22] |
Y. Yue, F. Huo, P. Yue, et al., Anal. Chem. 90 (2018) 7018-7024. DOI:10.1021/acs.analchem.8b01406 |
| [23] |
D.S. Leeman, K. Hebestreit, T. Ruetz, et al., Science 359 (2018) 1277-1283. DOI:10.1126/science.aag3048 |
| [24] |
G. Habicht, C. Haupt, R.P. Friedrich, et al., Proc. Natl. Acad. Sci. U. S. A. 104 (2007) 19232-19237. DOI:10.1073/pnas.0703793104 |
| [25] |
G.C.P. van Zundert, J.P.G.L.M. Rodrigues, M. Trellet, et al., J. Mol. Biol. 428 (2015) 720-725. DOI:10.1016/j.jmb.2015.09.014 |
| [26] |
D. Zhao, X. Dong, N. Jiang, et al., Nucleic Acids Res. 42 (2014) 11612-11622. DOI:10.1093/nar/gku833 |
| [27] |
S. Stefansson, D.L. Adams, C.M. Tang, et al., BioTechniques 52 (2012) 1-6. DOI:10.1155/2012/371487 |
| [28] |
J. Choi, S. Kim, T. Tachikawa, et al., J. Am. Chem. Soc. 133 (2011) 16146-16153. DOI:10.1021/ja2061984 |
| [29] |
A. Rajendran, S.I. Nakano, N. Sugimoto, Chem. Commun. 46 (2010) 1299-1301. DOI:10.1039/b922050j |
| [30] |
S. Pramanik, S. Nagatoishi, N. Sugimoto, Chem. Commun. 48 (2012) 4815-4817. DOI:10.1039/c2cc30622k |
| [31] |
R. Jin, B.L. Gaffney, C. Wang, et al., Proc. Natl. Acad. Sci. U. S. A. 89 (1992) 8832-8836. DOI:10.1073/pnas.89.18.8832 |
| [32] |
A. Rajendran, B.U. Nair, BBA-Gen, Subjects 1760 (2006) 1794-1801. DOI:10.1016/j.bbagen.2006.08.011 |
| [33] |
C.M. Karch, M. Prudencio, D.D. Winkler, et al., Proc. Natl. Acad. Sci. U. S. A. 106 (2009) 7774-7779. DOI:10.1073/pnas.0902505106 |
| [34] |
M. Chattopadhyay, A. Durazo, S.H. Sohn, et al., Proc. Natl. Acad. Sci. U. S. A. 105 (2008) 18663-18668. DOI:10.1073/pnas.0807058105 |
| [35] |
Z.A.O. Durer, J.A. Cohlberg, P. Dinh, et al., PLoS One 4 (2009) e5004. DOI:10.1371/journal.pone.0005004 |
| [36] |
J.S. Elam, A.B. Taylor, R. Strange, et al., Nat. Struct. Mol. Biol. 10 (2003) 461-467. DOI:10.1038/nsb935 |
| [37] |
M. Bucciantini, E. Giannoni, F. Chiti, et al., Nature 416 (2002) 507-511. DOI:10.1038/416507a |
| [38] |
I. Choi, Y.I. Yang, H.D. Song, et al., Biochim. Biophys. Acta 1812 (2011) 41-48. DOI:10.1016/j.bbadis.2010.09.003 |
| [39] |
S.J. Weisberg, R. Lyakhovetsky, A.C. Werdiger, et al., Proc. Natl. Acad. Sci. U. S. A. 109 (2012) 15811-15816. DOI:10.1073/pnas.1205829109 |
| [40] |
G.W. Collie, R. Promontorio, S.M. Hampel, et al., J. Am. Chem. Soc. 134 (2012) 2723-2731. DOI:10.1021/ja2102423 |
 2021, Vol. 32
2021, Vol. 32 

