Exosomes, which are secreted by most cell types and ranged in the size from 30 nm to 150 nm in diameter, play critical roles in various homeostatic processes, such as immune responses, viral pathogenicity, pregnancy, cells communication and cancer progression [1, 2]. A lot of research show that exosomes carry a broad cargo of proteins, metabolites, miRNA, and DNA derived from their parent cells [3] and this content is remarkably disease-specific [4]. In addition, exosomes are widely found in biological fluids, such as serum, saliva and urine. Given their unique characteristics, exosomes have received particularly attractive as minimally invasive liquid biopsies for following disease progression and treatment [5, 6]. Therefore, developing efficient methods for exosomes detection with high sensitivity and specificity is highly significant.
To date, many analytical methods have been developed to detect exosomes, including nanoparticle tracking analysis (NTA) [7, 8], western blot [9], ELISA [10, 11], and flow cytometry [12, 13], fluorescence [14, 15], colorimetry [16, 17], electrochemistry [18, 19], and surface enhanced Raman scattering (SERS) [20, 21]. Although most of these methods have been successfully applied to the study of exosomes, there are still some unresolved limitations. For example, NTA is non-specific and can only detect exosomes with a high concentration of 107–109 particles/mL [22]. Flow cytometry requires large amounts of sample input and the diameter of exosomes far below the detection limit of conventional flow cytometry, resulting in poor sensitivity and accuracy [12]. Moreover, since there are limited amounts of tumor-derived exosomes available during the early stages of cancer, these conventional exosomes detection methods are unable to distinguish between patients and healthy people. Therefore, more efficient methods need to be designed for exosomes sensing with high sensitivity and specificity.
Aptazyme, which contains both aptamer domain and DNAzyme domain, can simultaneously recognize specific targets and catalyze multiple-turnover reactions for signal generation and amplification [23-26]. This bifunctional feature makes aptazyme-based assays have attracted increasing attention for bioassay in recent years. For example, Li et al. reported a colorimetric ATP assay by using aptazyme-coupled rolling circle amplification [27]. Lu et al. developed an aptazyme-based label-free fluorescent probe for Pb2+ and adenosine detection [28]. By assembling aptazymes on the surface of gold nanoparticles, Wang et al. were able to utilize aptazymes for ATP imaging in living cells [29]. However, despite the significant efforts in developing aptazyme-based probes, there are no studies that have focused on applying this technology to exosome sensing.
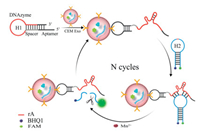
Herein, we report an aptazyme-based fluorescence probe for the amplified detection of exosomes. As illustrated in Scheme 1, the probe comprises of two components: i) A hairpin aptazyme (H1), consisting of a target-recognition aptamer sequence and a Mn2+-specific DNAzyme sequence; ii) a molecular beacon (H2), which is a substrate strand with a cleavage RNA site. The sequences of oligonucleotides involved are listed in Table S1 (Supporting information), the predicted secondary structure simulation diagram of H1 and H2 were provided in Fig. S1 (Supporting information). In this work, we chose the sgc8c aptamer as the basic model which can specifically target the protein tyrosine kinase-7 (PTK7) on the surface of CCRF-CEM cell [30]. In the presence of target exosomes, the specific binding between exosomes and aptamer in H1 induces a conformational transition, exposing the activated Mn2+-specific DNAzyme. The activated DNAzyme then cleaves the double labeled substrate H2 with the aid of Mn2+, resulting in the fluorescence restoration of FAM due to its separation from BHQ1. The released activated aptazyme can then bind to, and cleave another substrate strand. During this cyclic process, only a few exosomes are required to initiate the cleavage of multiple fluorophore-labeled substrate strands, which provides an amplified fluorescence signal and achieves enhanced sensitivity for detecting exosomes. Thanks to its high specificity and sensitivity, such an aptazyme-based approach is successfully used to detect exosomes in clinical serum samples from acute lymphoblastic leukemia patients (ALL), showing its tremendous potential for use in a clinical diagnosis.
|
Download:
|
| Scheme 1. Schematic diagram of the aptazyme-based fluorescence nanoprobe for ultrasensitive of exosomes via target-triggered DNAzyme cascade. | |
Exosome morphology was firstly characterized by scanning electron microscopy (SEM). As shown in Fig. S2A (Supporting information), exosomes have a uniform diameter with globoid vesicles between 30–100 nm, which is consistent with previous report [4]. The zeta potential of exosomes is −20.0±1.4 mV in HEPES buffer, which become −28.6±2.1 mV after the addition of H1 (Fig. S2B in Supporting information), suggesting the recognition and binding between aptamer and exosomes. The hydrodynamic diameter of exosomes recorded by dynamic light scattering (DLS) (Fig. S2C in Supporting information) is about 106.3±3.2 nm, which is consistent with the SEM result. In addition, the nanoparticle tracking analysis (NTA) assay determines that the exosomes have an average diameter of 139.5±1.1 nm and the corresponding concentration is 3.83×107 ± 3.11×106 particles/μL (Fig. S2D in Supporting information).
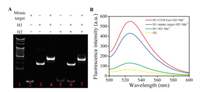
To evaluate the feasibility of our approach, the performance of the target-triggered DNAzyme cleavage was first investigated by native polyacrylamide gel electrophoresis used a mimic target (the complementary sequence of the aptamer in the aptazyme). As shown in Fig. 1A, the band of H2 in lane 7 disappear upon the addition of mimic target. But in the absence of mimic targets (lane 6), no obvious change can be seen in the H2 band. This result demonstrated that the mimic target could trigger the structural transformation of the aptazyme to activate the DNAzyme and cleave the substrate.
|
Download:
|
| Fig. 1. (A) Native PAGE gel assay. Lane 1: the standard DNA ladder; lane 2: mimic target; lane 3: H1; lane 4: mimic target + H1; lane 5: H2; lane 6: H1+H2 + Mn2+; lane 7: H1 + mimic target + H2 + Mn2+. (B) Fluorescence emission spectra of H2 (yellow curve), H1+H2 + Mn2+ (green curve), H1 + mimic target + H2 + Mn2+ (blue curve) and H1+H2 + Exo+Mn2+ (red curve). H1, H2 and mimic target are 100 nmol/L, Mn2+ is 500 μmol/L, and Exo is 3×105 particles/μL. | |
The fluorescence intensity of the system was also used to investigate the feasibility of the proposed assay. The exosome was first incubated with 10 nmol/L H1 in 85 μL of HEPES buffer at 37 ℃ for 2h. Then, 100 nmol/L H2 and 500 μmol/L Mn2+ were added and the mixture were incubated at 37 ℃ for another 2 h with a total volume of 100 μL. The fluorescence intensity, with an excitation wavelength of 488 nm, was measured. As seen in Fig. 1B, there is only a weak fluorescence signal in the absence of target exosomes (green curve). However, after introducing target exosomes, an obvious fluorescence increase can be observed (red curve), which is slightly higher than that caused by the mimic target (blue curve). The fluorescence recovery results demonstrated that the DNAzyme has been activated and the fluorescence substrate H2 was successfully cleaved. All these results indicated that this nanoprobe could be applied for exosomes sensing.
To obtain the best conditions for exosome detection, some important experimental parameters, including the sequence of H1, the ratio of H1 and H2, the concentration of Mn2+, reaction temperature and time were carefully investigated (Figs. S3–S7 in Supporting information). Using optimized conditions, this nanoprobe was first used to detect exosomes in buffer. As shown in Fig. 2A, the fluorescence intensity increased obviously with the increase of exosome concentration from 5×103 particles/μL to 2×106 particles/μL. The relationship between fluorescence intensity and exosome concentration is shown in Fig. 2B, where fluorescence intensity increases linearly with exosome concentrations from 5×103 particles/μL to 9×105 particles/μL. The correlation equation is fluorescence intensity=3.030[Exo] + 49.25 (R2=0.9987). According to the 3σ and 10σ rules, the LOD and LOQ for exosomes is estimated to be 3.5×103 particles/μL and 1.7×104 particles/μL, respectively, which is comparable to the results of previous studies (Table S2 in Supporting information).

|
Download:
|
| Fig. 2. (A) Fluorescence emission spectra of probe with different concentrations of exosomes: (a) 0, (b) 5×103, (c) 8×103, (d) 1×104, (e) 5×104, (f) 1×105, (g) 2×105, (h) 3×105, (i) 5×105, (j) 7×105, (k) 9×105, (l) 1.4×106, (m) 2×106 particles/μL. (B) Scatter plot of fluorescence intensity corresponding to (a) (0-2×106 particles/μL). Inset: The linear relationship between fluorescence intensity and concentration of exosomes ranging from 5×103 particles/μL to 9×105 particles/μL. | |
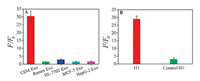
Specificity is another key issue that needs to be addressed when developed a new strategy. We therefore assessed the ability of this nanoprobe to distinguish among exosomes-derived from different cell lines. According to previous studies, PTK-7 is expressed on CCRF-CEM cells with a much higher level than that in other cell lines, such as Ramos and HL-7702 [31]. Therefore, specific experiments were conducted on exosomes derived form CCRF-CEM cells and other control cell lines with the same concentration. As shown in Fig. 3A, a significant fluorescence enhancement only can be seen in CCRF-CEM-derived exosomes, but there is no obvious fluorescence response for those derived from other lines including Ramos, HL-7702, MCF-7, and HepG-2. Meanwhile, a random sequence was chosen to replace the aptamer in H1 as a control. The result shows that the fluorescence intensity is almost the same as that in the blank (Fig. 3B). These all results demonstrated that the aptazyme based strategy possessed good selectivity due to its high specificity of aptamer in aptazyme.
|
Download:
|
| Fig. 3. (A) The response of nanoprobe to exosomes derived from CEM cells, Ramos cells, HL-7702 cells, MCF-7 cells, HepG-2 cells. (B) The fluorescence intensity changes of H1 and random control sequence after incubation with exosomes. The error bars represent the standard deviations of three repetitive measurements. | |
To investigate the anti-interference ability of our nanoprobe in sensing tumor-related exosomes, the CCRF-CEM cell-derived exosomes was first quantified in fetal bovine serum (FBS). Because FBS contains a number of components with high concentration, ultracentrifuged (UC) FBS was used as a mimetic clinical sample to evaluate the performance of exosome probe. Purified exosomes derived from CCRF-CEM cells were spiked in 0, 5%, 10%, 20%, 30% and 40% (v/v) UC FBS. As shown in Fig. S8 (Supporting information), the results obtained from UC FBS were almost the same as that in buffer. These results indicated that the system has high anti-interference ability for the detection of CCRF-CEM exosomes in simulated biological samples.
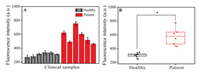
The exosome detection strategy was further applied to clinical blood samples from healthy donors and patients with acute lymphoblastic leukemia. The serum samples were provided by the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China). All experiments were performed in accordance with the Guidelines of Clinical Sample Management Rules of the First Affiliated Hospital of Zhengzhou University, and approved by the Ethics Committee at the First Affiliated Hospital of Zhengzhou University. Informed consent was obtained from the blood donors of this project. All patients were diagnosed, and their clinical test results confirmed as positive by using a blood screening test after displaying specific clinical symptoms. In the healthy donors, physical examination confirmed all range indicators used were in the normal value for test results. The procedure for exosomes detection in real human serum samples was the same as that in buffer. As shown in Figs. 4A and B, the fluorescence intensity of FAM in the acute lymphoblastic leukemia patient samples was consistently higher than that in the healthy group, demonstrating that this assay can distinguish between patient and healthy control samples.
|
Download:
|
| Fig. 4. (A) Histogram and (B) box plots result of exosomes detection in the clinical samples (*P < 0.05). | |
To further demonstrate the practical application of the aptazyme based probe, the NTA was introduced as a control to study the accuracy of this method. ExoEasy was first used to isolate exosomes from ten freshly collected clinical serum samples and then quantified them by NTA. Subsequently, the samples were assayed by our designed probe. As shown in Fig. S9 (Supporting information), the data obtained by our sensing method was consistent with the NTA data. Furthermore, the applicability of the aptazyme-based probe was also evaluated by recording the recoveries of exosomes in 1% diluted healthy serum samples. The results show a good agreement with the added concentration with recoveries from 96.0% to 103.0% (Table 1). These results indicated that our assay could work well in clinical samples and has great potential for application in clinical settings.
|
|
Table 1 Recovery results of exosomes spiked in 1% diluted clinical serum samples. |
In summary, by employing the advantages of aptazyme with high specificity and amplified ability, we developed an ultrasensitive and specific strategy for detecting cancer-related exosomes. This aptazyme-based assay possesses several advantages over currently reported methods. First, the use of aptazyme provides higher recognition efficiency. Second, the recognition trigged the activated of DNAzyme make the system possess higher signal to background, resulting in high sensitivity. Under optimized conditions, the assay has an excellent detection range from 5×103 particles/μL to 2×106 particles/μL with the LOD and LOQ as low as 3.5×103 particles/μL and 1.7×104 particles/μL, respectively. Third, the combination of aptamer and DNAzyme produces an efficient and simple assay for distinguishing cancer patients form healthy individuals. Therefore, we envision that this nucleic acid-based cyclic amplification method could be implemented into the clinical workflow for non-invasive and routine early cancer screening and cancer recurrence monitoring. Moreover, this aptazyme-based nanoprobe may also attain diagnostic values in other diseases, for instance, infectious disease, central nervous system-related diseases and tuberculosis treatment.
Declaration of competing interestThe authors report no declarations of interest.
AcknowledgmentsThis work was supported by National Natural Science Foundation of China (Nos. 21605038 and 21974125), China Postdoctoral Science Foundation (No. 2019T120623).
Appendix A. Supplementary dataSupplementarymaterial related to this article canbefound, in the online version, at doi: https://doi.org/10.1016/j.cclet.2020.11.031.
| [1] |
P. Zhang, M. He, Y. Zeng, Lab Chip 16 (2016) 3033-3042. DOI:10.1039/C6LC00279J |
| [2] |
H. Xie, K. Di, R. Huang, et al., Chin. Chem. Lett. 31 (2020) 1737-1745. DOI:10.1016/j.cclet.2020.02.049 |
| [3] |
T. Pisitkun, R.F. Shen, M.A. Knepper, Proc. Natl. Acad. Sci. 101 (2004) 13368-13373. DOI:10.1073/pnas.0403453101 |
| [4] |
S.A. Melo, L.B. Luecke, C. Kahlert, et al., Nature 523 (2015) 177-182. DOI:10.1038/nature14581 |
| [5] |
J. Zou, M. Shi, X. Liu, et al., Anal. Chem. 91 (2019) 2425-2430. DOI:10.1021/acs.analchem.8b05204 |
| [6] |
B. Shao, Z. Xiao, Anal. Chim. Acta 1114 (2020) 74-84. DOI:10.1016/j.aca.2020.02.041 |
| [7] |
R.A. Dragovic, C. Gardiner, A.S. Brooks, et al., Nanomedicine Nanotech. Biol. Med. 7 (2011) 780-788. DOI:10.1016/j.nano.2011.04.003 |
| [8] |
E. van der Pol, F.A.W. Coumans, A.E. Grootemaat, et al., J. Thromb. Haemost. 12 (2014) 1182-1192. DOI:10.1111/jth.12602 |
| [9] |
H. Zhou, P.S.T. Yuen, T. Pisitkun, et al., Kidney Int. 69 (2006) 1471-1476. DOI:10.1038/sj.ki.5000273 |
| [10] |
M. Oliveira-Rodríguez, S. López-Cobo, H.T. Reyburn, et al., J. Extracell. Vesicles 5 (2016) 31803-31812. DOI:10.3402/jev.v5.31803 |
| [11] |
P.G. Moon, J.E. Lee, Y.E. Cho, et al., Clin. Cancer Res. 22 (2016) 1757-1766. DOI:10.1158/1078-0432.CCR-15-0654 |
| [12] |
V. Pospichalova, J. Svoboda, Z. Dave, et al., J. Extracell. Vesicles 4 (2015) 25530-25544. DOI:10.3402/jev.v4.25530 |
| [13] |
W. Shen, K. Guo, G.B. Adkins, et al., Angew. Chem. Int. Ed. 57 (2018) 15675-15680. DOI:10.1002/anie.201806901 |
| [14] |
J. Chen, H.M. Meng, Y. An, et al., Talanta 209 (2020) 120510. DOI:10.1016/j.talanta.2019.120510 |
| [15] |
J. Chen, J. Tang, H.M. Meng, et al., Chem. Commun. 56 (2020) 9024-9027. DOI:10.1039/D0CC02337J |
| [16] |
Y. Xia, M. Liu, L. Wang, et al., Biosens. Bioelectron. 92 (2017) 8-15. DOI:10.1016/j.bios.2017.01.063 |
| [17] |
Z. Huang, Q. Lin, X. Ye, et al., Talanta 218 (2020) 121089. DOI:10.1016/j.talanta.2020.121089 |
| [18] |
W. Cheng, J. Ma, P. Cao, et al., Talanta 218 (2020) 121242. |
| [19] |
Y. Cao, L. Li, B. Han, et al., Biosens. Bioelectron. 141 (2019) 111397. DOI:10.1016/j.bios.2019.111397 |
| [20] |
T.D. Li, R. Zhang, H. Chen, et al., Chem. Sci. 9 (2018) 5372-5382. DOI:10.1039/C8SC01611A |
| [21] |
D. Ma, C. Huang, J. Zheng, et al., Biosens. Bioelectron. 101 (2018) 167-173. DOI:10.1016/j.bios.2017.08.062 |
| [22] |
E.I. Buzás, C. Gardiner, C. Lee, et al., Platelets 28 (2017) 249-255. DOI:10.1080/09537104.2016.1260704 |
| [23] |
X.X. Meng, X.H. Yang, K.M. Wang, et al., Chin. Chem. Lett. 20 (2009) 990-994. DOI:10.1016/j.cclet.2009.03.028 |
| [24] |
M. Wang, D. Yue, Q. Qiao, et al., Chin. Chem. Lett. 29 (2018) 703-706. DOI:10.1016/j.cclet.2018.03.025 |
| [25] |
Z. Zhang, A. Runa, J. Wu, et al., Chin. Chem. Lett. 30 (2019) 779-782. DOI:10.1016/j.cclet.2018.10.019 |
| [26] |
J. Huang, Y. He, X.H. Yang, et al., Chin. Chem. Lett. 25 (2014) 1211-1214. DOI:10.1016/j.cclet.2014.05.039 |
| [27] |
M.M. Ali, Y. Li, Angew. Chem. Int. Ed. 48 (2009) 3512-3515. DOI:10.1002/anie.200805966 |
| [28] |
P. Song, Y. Xiang, H. Xing, et al., Anal. Chem. 84 (2012) 2916-2922. DOI:10.1021/ac203488p |
| [29] |
Y. Yang, J. Huang, X. Yang, et al., Anal. Chem. 88 (2016) 5981-5987. DOI:10.1021/acs.analchem.6b00999 |
| [30] |
D. Shangguan, Z. Cao, L. Meng, et al., J. Proteome Res. 7 (2008) 2133-2139. DOI:10.1021/pr700894d |
| [31] |
W. Zhao, C.H. Cui, S. Bose, et al., Proc. Natl. Acad. Sci. 109 (2012) 19626-19631. DOI:10.1073/pnas.1211234109 |
 2021, Vol. 32
2021, Vol. 32 


