b School of Life Science, Advanced Research Institute of Multidisciplinary Science, Institute of Engineering Medicine, Key Laboratory of Molecular Medicine and Biotherapy, Beijing Institute of Technology, Beijing 100081, China
By regulating gene expression involved in the development and metastasis of cancer cells [1-4], RNA interference is demonstrated as a promising strategy for cancer treatment. Polycations, as one category of gene carriers, are generally applied to form polyelectrolyte complexes with siRNA for safe and efficient delivery [5-14]. Among the widely used polycations, peptides have received a great deal of attention because of their inherent biocompatibility and ease of manipulation [15-18]. Moreover, peptides with desirable sequences are functionalized and bioactive, which could further enhance the transfection efficacy [19-26].
siRNA carriers are generally administrated by intravenous injection. Their destinations in the blood vessels are controlled by at least two factors: the interaction with blood components and the deformation by blood flow. The particles with positive surface charge are subject to aggregation by interacting with the negatively charged proteins in serum [27-29]. The gene carriers also experience the shear force generated by blood flow. The shear rate of blood flow ranges from 20 s−1 to 2000 s−1 in healthy body [30, 31], which generates impacts on both the endogenous physiological activities and the exogenous drug delivery [31-35]. For example, the unfolding of von Willebrand factor induced by shear force is related to the platelet aggregation and thrombus formation [31, 35]. Our previous work has demonstrated that the flow shear affects polyelectrolyte complexes in two ways: stirring the system to make it more homogenous which may induce further aggregation, and tearing apart the particles at shear rate above certain value [36, 37].
Lung cancer has high incidence and is the leading cause of cancer-related mortality worldwide [38]. Because the particles administrated intravenously are firstly pumped by heart into the pulmonary capillary network before being distributed to the body [39, 40], the interaction with blood components and the deformation under blood flow could be applied to design siRNA carriers for targeted lung therapy. It is reported that micron-sized particles, especially those with size over 7μm in diameter, can rapidly accumulate in lung by mechanical filtration through capillary bed after intravenous administration [41-45]. This strategy, however, is limited because of the risk of undesirable side effects and even death due to long time blockade of blood vessels [41, 46]. To improve this situation, Couvreur et al. [47] designed some particles based on metal organic framework (MOF). The particles can accumulate in lung via aggregation, and disaggregate within 24h to release drug and avoid long time blockade. Inspired by these findings, we speculate that fabrication of siRNA delivery vectors that can interact with blood components to form large size aggregates, and can further break down by blood shear should be a promising approach to achieve safe and efficient lung-targeting gene delivery as it is self-regulated by circulatory system.
The aggregation of formulated siRNA particles with blood components has been well-studied in literature [48, 49]. However, the breakdown mechanism of the aggregates by blood shear has not been clarified, not mentioning its relationship with the targeting efficacy. Herein, we designed a series of peptides which can form complexes with siRNA and tested the behaviors of the complexes in blood stream. Since the surface property and the viscoelasticity of the complex are the key parameters affecting their response to serum and shear force, the peptides are designed with specific consideration on charge density, hydrophobicity, and secondary structures.
For a better comparison, all the artificial peptides are fixed with 33 amino acid residues and equal charge density (Table 1), so that they have the same capacity to form complexes with siRNA via electrostatic interactions. The sequence can be divided into two parts linked by three glycine residues. The arginine-rich sequence can condense siRNA to form complex. The hydrophobicity of the peptides is tuned by the number of incorporated leucine or serine residues. The peptide named RL contains hydrophobic leucine (L) residues, while the peptide named RS contains hydrophilic serine (S) residues. The peptides with intermediate hydrophobicity are named as RLSb and RLSa, with the L and S residues in block arrangement and in alternative arrangement, respectively. The (FKFE)3 sequence, which is able to form inter-chain β-sheet structure [50-52] and functions as dynamic cross-linking point, is introduced as the second part in the peptide to stabilize the siRNA complex. A control peptide (RS-Nβ) without (FKFE)3 is also designed to reveal the effect of β-sheet structure.
|
|
Table 1 Name and sequence of designed peptides. |
In order to investigate the differences among complexes resulting from peptides sequence, the effect of other parameters, including mixing ratio, concentration, order of mixing, ionic strength, pH should be eliminated. On the basis of our previous study [53, 54] and the findings on siRNA delivery in literature [55, 56], siRNA with the concentration 5 times higher is added to peptide in DPBS buffer to ensure that the final peptide/siRNA mixing charge ratio is 20 and the dilution effect is alleviated. This charge ratio also ensures that all siRNA molecules are incorporated inside the complexes. DPBS buffer fixes the ionic strength and pH value. FBS is added to the complexes to mimic the blood environment. Because the total concentration of protein in FBS is about 35−45 mg/mL, several orders higher than those of peptides and siRNA, and conceals the behaviors of complexes in the mixture, the FBS concentration is diluted to 5 vol%. Even though the concentration of protein in 5 vol% FBS is still higher than those of peptides and siRNA, laser light scattering (LLS), which is much more sensitive to particle size than concentration, is able to detect the peptide/siRNA complexes in serum under such conditions.
The behavior of complexes in blood environment depends on both inherent structure of complexes and the interaction between peptides and serum components (Fig. 1). As concentrated siRNA solution is added to peptide solution to reach the +/− ratio of 20, complex is formed instantly in all the studied cases. The stability of these complexes is peptide dependent (Figs. 1a1-e1 and Fig. S1 in Supporting information). The complex formed by RS and siRNA is the most unstable one, whose size and excess scatted intensity drop sharply and reach the values of individual peptide in 90min (Fig. 1d1 and Fig. S1b). On the contrary, the complex formed by RL and siRNA is much more stable. The excess scattered intensity slightly declines with time (Fig. S1b), and the average size does not show prominent change, except for a broadening in size distribution in 3h (Fig. 1a1). RLSa and RLSb only differ in sequence, but their complexes with siRNA exhibit huge difference. The RLSa/siRNA complex has similar scattered intensity and size as the RL/siRNA complex, and is even more stable under the same conditions (Fig. 1c1 and Fig. S1b). The RLSb/siRNA complex, however, is smaller in size (130 nm) (Fig. 1b1) and much lower in scattered intensity, which also decreases with time (Fig. S1b). The RS-Nβ/siRNA complex does not contain β-sheet structures. The Rh, app value is extremely large and increases with time, while the excess scattered intensity is low and constant (Fig. 1e1 and Fig. S1b). The chain density of the complex, which is proportional to Iex/R3 [57], should be much lower than that of the complex containing β-sheet structures.

|
Download:
|
| Fig. 1. Time dependence of size distribution of peptides/siRNA (a1-e1), peptides/FBS (a2-e2) and peptides/siRNA/FBS (a3-e3), respectively. Scattering angle: 30°. | |
5 vol% FBS exhibits a broad distribution in DPBS buffer [37], because of the existence of multiple components. The excess scattered intensity of 5 vol% FBS is also higher than those of the peptides at 0.1 mg/mL (Fig. S1a). 10 vol% FBS is added into 0.1 mg/mL peptides with equal volume, and the final concentration of FBS is 5 vol%. When the peptides are mixed with FBS, the size distribution (Figs. 1a2-e2) and the increase of the excess scattered intensity (Fig. S1c) indicate the occurrence of aggregation in all the studied cases. The degree of aggregation is peptide dependent. The aggregates formed by hydrophilic peptide RS and serum components are at least three times larger in Rh, app (170 nm, Fig. 1d2) and thirty time higher in excess scattered intensity (Fig. S1c) compared with the aggregates formed by hydrophobic peptide RL and the serum components under the same conditions. RLSa in 5 vol% FBS is similar to RS, while RLSb is closer to RL (Fig. 1 and Fig. S1c). The behavior of RS-Nβ is quite different from that of RS. It forms an aggregate with Rh, app more than 100 nm after one hour in 5 vol% FBS (Fig. 1e2), while the excess scattered intensity is much lower (Fig. S1c), indicating that the aggregates of RS-Nβ/FBS contain loose structures.
The peptide/siRNA complex at the +/− ratio of 20 is mixed with equal volume of 10 vol% FBS 30min after preparation, so that the final concentration of FBS is 5 vol%. The complex with excess peptides on the surface undergo further aggregates in 5 vol% FBS (Figs. 1a3-e3 and Fig. S1d). The complexes formed by siRNA and the peptides containing β-strands exhibit mainly monomodal distribution in 5 vol% FBS (Figs. 1a3-d3), and the average sizes are around 270 nm. However, the excess scattered intensity of the complexes varies with the peptides. The RS/siRNA complex exhibits the highest excess scattered intensity in 5 vol% FBS (Fig. S1d). The excess scattered intensities of the rest complexes follow the order of RLSa > RLS b ≅ RL (Fig. S1d). The RS-Nβ/siRNA complex in 5 vol% FBS is also different from the others. A multimodal distribution with the size of the largest aggregate reaching microns is observed (Fig. 1e3). Again, the excess scattered intensity is the lowest in all the studied cases (Fig. S1d).
The peptide/siRNA/FBS complexes are treated by shear at different shear rates for 2h to reveal the effect of blood flow. Because the blood shear rate covers a broad range and varies with vessels [30] as well as the blockage of the vessels, we choose 100~1000 s−1 to study the effect of shear rates. Our previous studies [36, 37] have demonstrated that the shear force not only has the stirring effect to facilitate the interaction between the particle and the environment, it is also able to split the particle into smaller pieces as the shear rate reaches a critical value. The critical value is related to size, viscoelasticity, and other intrinsic properties of the particles. The peptide/siRNA/FBS complex is not uniform. It contains multiple domains different in charge, density, and viscoelasticity. The excess FBS components in the environment can further interact with the complex as it is deformed or split under shear. Therefore, the overall shear effects are complicated but peptide-related. The behaviors of RL/siRNA/FBS and RLSb/siRNA/FBS are similar. At lower shear rate such as 100 s−1, a further aggregation of the complexes as demonstrated by the increase in size, as well as a spinoff of particles as demonstrated by the appearance of a small size peak (Figs. 2a and b), are observed, suggesting that the heterogeneous peptide/siRNA/FBS complexes experience both the shear effects simultaneously. The loose periphery of the complex is liable to split under shear, while the exposure of the complex interior results in enhanced aggregation with FBS. The former leads to a decrease in the excess scattered intensity, while the latter shows the opposite trend. The overall change of scattered intensity is determined by the balance of the two effects (Fig. S2 in Supporting information). With increasing shear rate to 500 s−1 or above, a further decrease in both the aggregate size (250 nm at 500 s−1 and 210 nm at 1000 s−1 for RL/siRNA/FBS, 270 nm at 500 s−1 and 230 nm at 1000 s−1 for RLSb /siRNA/FBS) and the excess scattered intensity is observed (Fig. S2), suggesting that the spinoff of smaller size particles is the dominant process. Likewise, both larger aggregates and smaller pieces are observed as RS/siRNA/FBS is treated at 100 s-1 (Fig. 2d). The size of the aggregates and the scattered light intensity also decrease with increasing shear rate. However, the sizes of the aggregate of RS/siRNA/FBS are 380 nm at 500 s−1 and 260 nm at 1000 s−1, significantly larger than those of the aggregates formed RL/siRNA/FBS and RLSb/siRNA/FBS under the same conditions (Fig. 2). The complexes of RLSa/siRNA/FBS and RS-Nβ/siRNA/FBS are special. The size of RLSa/siRNA/FBS does not show prominent change until the shear rate reaches 1000 s−1 (Fig. 2c). The RS-Nβ/siRNA/FBS complex exhibits a multiple modal distribution with the largest peak in the order of micron (Fig. 2e), and the excess scattered intensity is the lowest in all the complexes (Fig. S2). With increasing shear rate from 100 s−1 to 1000 s−1, the multimodal distribution evolves into a bimodal distribution with the Rh, app of the two fragments being 310 nm and 28 nm, respectively (Fig. 2e). Meantime, the excess scattered intensity increases by a factor of 5 (Fig. S2), suggesting that the resulting RS-Nβ/siRNA/FBS complex after shear treatment possesses higher chain density. The morphologies of peptide/siRNA/FBS are also revealed by AFM experiment. The complexes exhibit mainly spheres or cluster of spheres after being dried on the mica surface (Fig. S3 in Supporting information). The variation of size distribution at different shear rate observed in AFM agree with LLS result.

|
Download:
|
| Fig. 2. Size distribution of peptides/siRNA/FBS before (a0-e0) after being sheared at 100 s−1 (a1-e1), 500 s−1 (a2-e2), 1000 s−1 (a3-e3), separately, for 2 h. Scattering angle: 30°. Gray dot line: 480 nm. | |
Peptide/siRNA complexes at the +/− ratio of 20 are administered in C57BL/6 mice via tail vein to test the in vivo biodistribution. The siRNA dose is 1.0 mg/kg on the basis of the mouse weight. Naked Cy5-labeled siRNA, corresponding to the dose in the complex, and PBS buffer are also administrated, separately, as controls. The tissue distribution of siRNAs is monitored over a course of 48 h using an in vivo imaging system. Compared to naked-siRNA, peptide-formulated siRNA presents a significantly prolonged circulation time, and a distinct tissue distribution profile (Fig. 3 and Fig. S4 in Supporting information). The naked-siRNA accumulates in liver or kidney after intravenously administrated, while the peptide-formulated siRNA exhibits extremely high signal intensity in lung. Unfortunately, the mice injected with RS/siRNA and RS-Nβ/siRNA are all died with bleeding in nose and mouth after 1 h and 6 h, respectively. Postmortem reveals hyperemia and hemorrhage in lungs of these mice, indicating the occurrence of pulmonary embolism. Interestingly, the mice injected with RLSa/siRNA are died immediately after administration. Autopsy results show no prominent pulmonary embolism. We suspect the death could be related to the sequence of RLSa. Detailed study on this issue is beyond the scope of the current work.

|
Download:
|
| Fig. 3. In vivo distribution of peptide/siRNA complexes. (a) Whole-body imaging, three mice are included in each treatment group. The mice injected with RS/siRNA and RS-Nβ/siRNA are died in 1 h and 6 h, respectively. 1.5 h and 48 h post administration, siRNA accumulation in the major tissues is visually examined (b, c). PBS, mice treated only with saline; naked-siRNA, mice treated with free siRNA without any carrier. | |
The mice survive after the intravenously administration of both RL/siRNA complex and RLSb/siRNA complex. The signal intensity (Fig. 3c) shows that the accumulation of RL/siRNA complex in lung 1.5 h after administration is much higher than that in other tissues, such as liver, kidney, and spleen. The trend remains till 48 h. The RLSb/siRNA complex exhibits almost equal accumulation in lung, liver, and kidney in 1.5 h (Fig. 3b). However, the intensity in kidney is higher than that in lung after 48 h, suggesting that the RLSb/siRNA complexes tend to dissociate and pass through the pulmonary capillaries, exhibiting a behavior similar as naked siRNA.
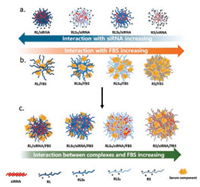
To explain the response of complexes to shear flow and biodistribution in vivo, the interactions other than electrostatic interaction, such as dynamic cross-linking, hydrophobic interaction, and hydrogen bonding, should also be considered. Fig. 1 demonstrates that RL has a stronger tendency to form complex with siRNA than RS, indicating that the hydrophobic interaction enhances and stabilizes the siRNA complexes. On the contrary, RS interacts with FBS much more strongly than RL (Fig. 1). We attribute it to the hydrogen bonds formed by serine residues in RS with the components in FBS. RLSa and RLSb contain the same components, but differ in sequence. The former exhibits higher capacity than the latter in forming complexes with both siRNA and FBS (Fig. 1 and Fig. S1), suggesting that even distribution of leucine residues and serine residues along the peptide backbone (RLSa) yields both stronger hydrophobic interaction with siRNA and hydrogen bonding with serum components than the distribution of corresponding residues in block formation (RLSb). Since the peptide/siRNA complexes at the +/− ratio of 20 contain a large amount of excess peptides, the interaction of the complexes with FBS (Figs. 1a3-e3 and Fig. S1d) follows a similar trend as that of the individual peptide interacting with FBS (Figs. 1a2-e2 and Fig. S1c). The trends of the peptides interacting with siRNA and FBS are schematically shown in Fig. 4. The β-sheet serves as dynamic cross-linking points, which has the capacity to stabilize and tighten the complexes into more compact form. The peptide/siRNA/FBS complexes in Fig. 4 contain distinct domains, and each domain possesses its specific response to shear, the peptide/siRNA/FBS complexes with multiple domains exhibit peptide-dependent profiles with increasing shear rate. The behavior of RL/siRNA/FBS complex is similar to other core-corona complexes under shear [37, 58]: the corona is firstly distorted at lower shear rate to expose the RL wrapped inside, which leads to a further aggregation via the interaction between exposed RL and free FBS in the solution. With increasing shear rate above 100 s−1, the aggregates break down into small size fragments (Fig. 2a). The RL/siRNA complexes can effectively accumulate in the lung of mice without causing prominent pulmonary embolism, indicating that the blood flow in the lung is suitable to breakdown the RL/siRNA/FBS complex particles.
|
Download:
|
| Fig. 4. Schematically showing the interaction of peptides with siRNA (a), peptides with FBS (b), and the peptide/siRNA complexes with FBS (c). | |
The RLSb/siRNA/FBS complex exhibits similar behavior as the RL/siRNA/FBS complex. RLSb has the same charge density as RL, but less number of hydrophobic leucine residues. Therefore, the RLSb/siRNA complex core is not as dense and stable as the RL/siRNA complex core (Fig. 1b1 and Fig. S1b). Most RLSb/siRNA/FBS complex is prone to break into small particles and go through the capillaries of lung, which would decrease their targeting efficiency [44] (Fig. 2b). The in vivo test also confirms this conclusion (Fig. 3).
RS is even more hydrophilic than RLSb. It does not effectively form complex with siRNA since the complex dissociates with time (Fig. 1d1 and Fig. S1b). However, RS interact strongly with FBS via both electrostatic interaction and hydrogen bonds, which controls the formation of RS/siRNA/FBS complexes. As the complex breaks down into small size fragments, the exposed RS can further form aggregates with the FBS in the environment. At high shear rate, there still exist large size RS/siRNA/FBS particles (Fig. 2d), as demonstrated by the relatively higher scattered intensity (Fig. S2). When the particles are injected intravenously, pulmonary embolism is inevitable.
Even though RLSa and RLSb only differ in sequence, the even distribution of serine and leucine residues along the backbone renders RLSa both strong hydrophobic interaction (similar as RL in Fig. 1c1) and hydrogen bonds (similar as RS in Fig. 1c2). Together with the electrostatic interaction, the RLSa/siRNA/FBS complex is tougher than other complexes under shear (Fig. 2c). RLSa/siRNA/FBS exhibits acute toxicity after being injected and causes sudden death of the mice. The toxicity could be attributed to certain sequence of RLSa, which is not clear at current stage. The RS-Nβ/siRNA/FBS complexes form clusters of huge sizes. Even though shear, functioned as a homogenizer, breaks down the clusters and enhances the chain density, the size of the clusters still large enough to cause pulmonary embolism, leading to the death of mice.
Fabrication of vectors sensitive to blood flow is an optimal approach to targeting siRNA to lung via self-regulation. The responses to shear and the interaction with FBS components are two additional yet crucial factors to be considered when designing peptide as the delivery vehicles. The peptides should interact more strongly with siRNA than with the FBS components so that the complex contains distinct domains or hierarchical structures: the dense peptide/siRNA domains prevent the complexes from being broken down beforehand and the loose peptide/FBS junctions break down by shear with ease to avoid pulmonary embolism. Peptide is probably the best candidate for the blood-regulated lung-targeting siRNA delivery vehicle because of its high programmability to tune the balance between different interactions, including electrostatic interaction, hydrogen bonding, hydrophobic interaction, and so on. The cooperation of different interactions may result in specific binding of the peptide to certain biomolecules or tissues, yielding unexpected effect. Designing peptides with less mount of amino acid residues and simple sequence could be a solution to avoid complication.
Declaration of competing interestThe authors report no declarations of interest.
AcknowledgmentsThis work was supported by Beijing Natural Science Foundation (No. 2171001) and the National Natural Science Foundation of China (Nos. 21973002, 21774002, 31871003, 31901053), the Beijing-Tianjin-Hebei Basic Research Cooperation Project (No. 19JCZDJC64100), the Beijing Nova Program from Beijing Municipal Science & Technology Commission (No. Z201100006820005), the Young Elite Scientist Sponsorship Program of Beijing Association for Science and Technology (No. 2020-2022). We thank Biological & Medical Engineering Core Facilities (Beijing Institute of Technology) for providing advanced equipment and help.
Appendix A. Supplementary dataSupplementary material related to this article can befound, in the online version, at doi: https://doi.org/10.1016/j.cclet.2020.12.005.
| [1] |
M.L. Bobbin, J.J. Rossi, Annu. Rev. Pharmacol. Toxicol. 56 (2016) 103-122. DOI:10.1146/annurev-pharmtox-010715-103633 |
| [2] |
Y. Weng, H. Xiao, J. Zhang, X.J. Liang, Y. Huang, Biotechnol. Adv. 37 (2019) 801-825. DOI:10.1016/j.biotechadv.2019.04.012 |
| [3] |
Y. Xiao, K. Shi, Y. Qu, B. Chu, Z. Qian, Mol. Ther. Methods Clin. Dev. 12 (2019) 1-18. DOI:10.1016/j.omtm.2018.09.002 |
| [4] |
R.L. Setten, J.J. Rossi, S.P. Han, Nat. Rev. Drug Discovery 18 (2019) 421-446. DOI:10.1038/s41573-019-0017-4 |
| [5] |
W. Ho, X.Q. Zhang, X.Y. Xu, Adv. Healthcare Mater. 5 (2016) 2715-2731. DOI:10.1002/adhm.201600418 |
| [6] |
B. Kim, J.H. Park, M.J. Sailor, Adv. Mater. 31 (2019) e1903637. DOI:10.1002/adma.201903637 |
| [7] |
H. Gao, H. Feng, Y. Bai, et al., J. Biomed. Nanotechnol. 15 (2019) 1764-1770. DOI:10.1166/jbn.2019.2805 |
| [8] |
J. Ma, J. Zhang, L. Chi, et al., Chin. Chem. Lett. 31 (2020) 1427-1431. DOI:10.1016/j.cclet.2020.02.034 |
| [9] |
T. Frohlich, E. Wagner, Soft Matter 6 (2010) 226-234. DOI:10.1039/B916053A |
| [10] |
Y.X. Chen, B.W. Li, X.H. Chen, et al., Chin. Chem. Lett. 31 (2020) 1153-1158. DOI:10.1016/j.cclet.2019.06.022 |
| [11] |
T.B. Zhang, Y.Y. Huang, X.W. Ma, et al., Nano Lett. 18 (2018) 6301-6311. DOI:10.1021/acs.nanolett.8b02553 |
| [12] |
X.X. Liu, J.H. Zhou, T.Z. Yu, et al., Angew. Chem. Int. Ed. 53 (2014) 11822-11827. DOI:10.1002/anie.201406764 |
| [13] |
Y.Q. Wang, C.J. Li, L. Du, Y. Liu, Chin. Chem. Lett. 31 (2020) 275-280. DOI:10.1016/j.cclet.2019.03.040 |
| [14] |
Y.Y. Huang, D.S. Lin, Q. Jiang, et al., Biomaterials 33 (2012) 4653-4664. DOI:10.1016/j.biomaterials.2012.02.052 |
| [15] |
R.S. Shukla, B. Qin, K. Cheng, Mol. Pharmaceutics 11 (2014) 3395-3408. DOI:10.1021/mp500426r |
| [16] |
K. Numata, Polym. J. 47 (2015) 537-545. DOI:10.1038/pj.2015.35 |
| [17] |
J.Y. Zhao, X.F. Jiang, Chin. Chem. Lett. 29 (2018) 1079-1087. DOI:10.1016/j.cclet.2018.05.026 |
| [18] |
J. Liu, N.N. Guo, C. Gao, et al., J. Biomed. Nanotechnol. 15 (2019) 531-543. DOI:10.1166/jbn.2019.2705 |
| [19] |
T. Singh, A.S.N. Murthy, H.J. Yang, J. Im, Drug Deliv. 25 (2018) 1996-2006. DOI:10.1080/10717544.2018.1543366 |
| [20] |
J.C. Cummings, H.W. Zhang, A. Jakymiw, Transl. Res. 214 (2019) 92-104. DOI:10.1016/j.trsl.2019.07.010 |
| [21] |
W.Y. Tai, X.H. Gao, Adv. Drug Delivery Rev. 110 (2017) 157-168. |
| [22] |
Y. Yang, Y.P. Jia, Y. Xiao, et al., ChemPhysChem 19 (2018) 2058-2069. DOI:10.1002/cphc.201701375 |
| [23] |
M.F. Zhang, W.Y. Lu, Acta Pharm. Sin. B 8 (2018) 106-115. DOI:10.1016/j.apsb.2017.11.004 |
| [24] |
R.F. Wang, Q. Shen, X. Li, et al., Acta Pharm. Sin. B 8 (2018) 825-832. DOI:10.1016/j.apsb.2018.06.006 |
| [25] |
Y.H. Li, K.A. Clark, Z.P. Tan, Chin. Chem. Lett. 29 (2018) 1074-1078. DOI:10.1016/j.cclet.2018.05.027 |
| [26] |
Q.Y. Kuai, Y. Wang, F.H. Gao, et al., J. Biomed. Nanotechnol. 15 (2019) 979-992. DOI:10.1166/jbn.2019.2764 |
| [27] |
A. Gessner, A. Lieske, B.R. Paulke, R.H. Muller, J. Biomed. Mater. Res. A. 65A (2003) 319-326. DOI:10.1002/jbm.a.10371 |
| [28] |
Z.M. Zhao, A. Ukidve, V. Krishnan, S. Mitragotri, Adv. Drug Delivery Rev. 143 (2019) 3-21. DOI:10.1016/j.addr.2019.01.002 |
| [29] |
M.J. Ernsting, M. Murakami, A. Roy, S.D. Li, J.Control.Release 172 (2013) 782-794. DOI:10.1016/j.jconrel.2013.09.013 |
| [30] |
L.F. Brass, S.L. Diamond, J. Thromb. Haemost. 14 (2016) 906-917. DOI:10.1111/jth.13280 |
| [31] |
L.D.C. Casa, D.H. Deaton, D.N. Ku, J. Vasc. Surg. 61 (2015) 1068-1080. DOI:10.1016/j.jvs.2014.12.050 |
| [32] |
N. Korin, M. Kanapathipillai, B.D. Matthews, et al., Science 337 (2012) 738-742. DOI:10.1126/science.1217815 |
| [33] |
S.Y. Lee, M. Ferrari, P. Decuzzi, Nanotechnology 20 (2009) 495101. DOI:10.1088/0957-4484/20/49/495101 |
| [34] |
C. Fillafer, G. Ratzinger, J. Neumann, et al., Lab Chip 9 (2009) 2782-2788. DOI:10.1039/b906006e |
| [35] |
L.D.C. Casa, D.N. Ku, Annu. Rev. Biomed. Eng. Vol 19 (2017) 415-433. DOI:10.1146/annurev-bioeng-071516-044539 |
| [36] |
H. Wen, Q.H. Yu, Y.D. Yin, et al., Biomacromolecules 18 (2017) 3252-3259. DOI:10.1021/acs.biomac.7b00900 |
| [37] |
H. Wen, D.X. Yin, G.Q. Wu, et al., Soft Matter 16 (2020) 2301. DOI:10.1039/C9SM02397F |
| [38] |
F. Bray, J. Ferlay, I. Soerjomataram, et al., CA: Cancer J. Clin. 68 (2018) 394-424. DOI:10.3322/caac.21492 |
| [39] |
R.M. Schwartzstein, M.J. Parker, Respiratory Physiology: A Clinical Approach, 1st ed., LWW, 2005.
|
| [40] |
G.A. Truskey, F. Yuan, D.F. Katz, Transport Phenomena in Biological System, 2nd ed., Pearson, 2009.
|
| [41] |
Y.M. Wei, L. Zhao, Pharm. Dev. Technol. 19 (2014) 129-136. DOI:10.3109/10837450.2012.757782 |
| [42] |
C. Dhand, M.P. Prabhakaran, R.W. Beuerman, etal., RSCAdv. 4 (2014) 32673-32689. |
| [43] |
J.Y. Park, S. Park, T.S. Lee, et al., Biomaterials 218 (2019) 119331. DOI:10.1016/j.biomaterials.2019.119331 |
| [44] |
H.L. Kutscher, P. Chao, M. Deshmukh, et al., J. Control.Release 143 (2010) 31-37. DOI:10.1016/j.jconrel.2009.12.019 |
| [45] |
N.M. Pinkerton, S.W. Zhang, R.L. Youngblood, et al., Biomacromolecules 15 (2014) 252-261. DOI:10.1021/bm4015232 |
| [46] |
A.E. Jones, J.A. Watts, J.P. Debelak, et al., Am. J. Physiol. Lung C 284 (2003) L1072-L1081. DOI:10.1152/ajplung.00283.2002 |
| [47] |
T. Simon-Yarza, M. Gimenez-Marques, R. Mrimi, et al., Angew. Chem. Int. Ed. 56 (2017) 15565-15569. DOI:10.1002/anie.201707346 |
| [48] |
A.M. O'Mahony, S. Desgranges, J. Ogier, et al., Pharm. Res. 30 (2013) 1086-1098. DOI:10.1007/s11095-012-0945-8 |
| [49] |
J.F. Guo, W.P. Cheng, J.X. Gu, et al., Eur. J. Pharm. Sci. 45 (2012) 521-532. DOI:10.1016/j.ejps.2011.11.024 |
| [50] |
C.J. Bowerman, B.L. Nilsson, Biopolymers 98 (2012) 169-184. DOI:10.1002/bip.22058 |
| [51] |
M.R. Caplan, E.M. Schwartzfarb, S.G. Zhang, R.D. Kamm, D.A. Lauffenburger, Biomaterials 23 (2002) 219-227. DOI:10.1016/S0142-9612(01)00099-0 |
| [52] |
M.R. Caplan, P.N. Moore, S.G. Zhang, R.D. Kamm, D.A. Lauffenburger, Biomacromolecules 1 (2000) 627-631. DOI:10.1021/bm005586w |
| [53] |
J.H. Zhou, D. Li, H. Wen, et al., Sci. Rep.-U.K. 6 (2016) 22731. DOI:10.1038/srep22731 |
| [54] |
H. Wen, Y.D. Yin, C. Huang, W. Pan, D.H. Liang, Sci. China Chem. 60 (2017) 130-135. DOI:10.1007/s11426-016-0094-8 |
| [55] |
S. Mehrotra, I. Lee, C. Chan, Acta Biomater. 5 (2009) 1474-1488. DOI:10.1016/j.actbio.2009.01.004 |
| [56] |
Y. Wu, W.W. Wang, Y.T. Chen, et al., Int. J. Nanomed. 5 (2010) 129-136. |
| [57] |
X.H. Wang, X.P. Qiu, C. Wu, Macromolecules 31 (1998) 2972-2976. DOI:10.1021/ma971873p |
| [58] |
W. Pan, D.X. Yin, H.R. Jing, et al., Chin. J. Polym. Sci. 37 (2019) 36-42. DOI:10.1007/s10118-018-2166-z |
 2021, Vol. 32
2021, Vol. 32 

