b Key Laboratory for Ultrafine Materials of Ministry of Education, Frontiers Science Center for Materiobiology and Dynamic Chemistry, Research Center for Biomedical Materials of Ministry of Education, School of Materials Science and Engineering, East China University of Science and Technology, Shanghai 200237, China
Polypeptides have diverse biological functions and applications in antimicrobial [1-5], drug and gene delivery [6-10], bioimaging [11, 12], tissue engineering [13] and biomedical applications [14-22]. Nevertheless, the application of polypeptides suffers from their easy enzymatic degradation, high price and difficulties in large-scale synthesis. Polypeptoids are explored as mimics of polypeptides because they not only have excellent stability against proteolysis [23], but also can mimic the functions and applications of polypeptides [24-28]. Polypeptoids can be synthesized from one-pot ring-opening polymerization of amino acid N-substituted N-carboxyanhydrides (NNCAs) [29-35]. However, the polymerization of NNCAs may be very slow utilizing the widely used amine initiators, especially for NNCAs bearing a bulky N-substitution group such as cyclohexyl. The n-hexylamine-initiated polymerization on cyclohexyl-NNCA takes at least 6 days even at a high reaction concentration of 1 mol/L (Fig. 1a).
|
Download:
|
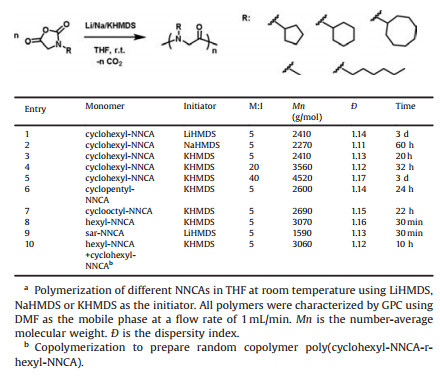
| Fig. 1. Inactive NNCA polymerization in THF. (a) Slow and fast cyclohexyl-NNCA polymerization initiated by n-hexylamine and KHMDS in THF at room temperature, respectively. (b) Remaining NNCA across the polymerization time. (c) GPC traces result of cyclohexyl-NNCA polymerization using DMF as the mobile phase at a flow rate of 1 mL/min. (d) GPC characterization result of cyclohexyl-NNCA polymerization. | |
Aforementioned challenges encourage us to explore new strategies for NNCAs polymerization. Recent studies in our lab demonstrated that lithium/sodium/potassium hexamethyldisilazide (Li/Na/KHMDS) all can initiate the superfast polymerization on amino acid N-carboxyanhydrides for easy synthesis of polypeptides [36-39], with KHMDS initiating the fastest polymerization. Our mechanism study indicates that Li/Na/KHMDS deprotonates the NH on the NCA ring and initiates the NCA polymerization, which implies naturally that Li/Na/KHMDS can initiate the ring-opening polymerization of NNCAs by deprotonation of the acidic CH on the NNCA ring. Experiments supported this hypothesis and demonstrated that KHMDS initiates the ring-opening polymerization of cyclohexyl-NNCA with a reaction rate much faster than does the n-hexylamine initiator and reduces the reaction time from 6 days to only 20 h, n-hexylamine vs. KHMDS as the initiator (Fig. 1a). When KHMDS-initiated cyclohexyl-NNCA completed after 20 h, only 12% of cyclohexyl-NNCA was consumed if using n-hexylamine as the initiator (Fig. 1b). It is also noteworthy that KHMDS-initiated polymerization also provided resulting polypeptoids with sharp signal of GPC trace and a narrow dispersity of 1.13 (Figs. 1c and d).
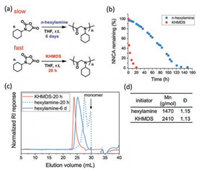
Our continuous study showed that Li/Na/KHMDS all can initiate the ring-opening polymerization of cyclohexyl-NNCA to give polypeptoids, with KHMDS initiated the fastest reaction (Table 1, entries 1–3). KHMDS can also initiate the polymerization of cyclohexyl-NNCA to provide polypeptoids with a higher molecular weight, Mn at 3560 and 4520 g/mol, but also with increased reaction time (entries 4 and 5, GPC traces in Fig. S16). KHMDS also successfully initiated the ring-opening polymerization of other inactive NNCAs with bulky N-substitution, such as cyclopentyl-NNCA and cyclooctyl-NNCA, to afford corresponding polypeptoids with a narrow dispersity (Đ=1.14−1.15; entries 6 and 7 in Table 1; GPC traces in Figs. 2a and b). It is noteworthy that for active NNCAs, such as hexyl-NNCA and sar-NNCA, a fast polymerization proceeded to afford corresponding polypeptoids within 30 min (entries 8 and 9 in Table 1; GPC trace in Figs. 2c and d). Moreover, copolymerization can also proceed successfully with this new NNCA polymerization strategy, as demonstrated by the synthesis of random copolymer poly(cyclohexyl-NNCA-r-hexyl-NNCA) using KHMDS as the initiator (entry 10 in Table 1; GPC trace in Fig. 2e).
|
|
Table 1 Alkali-metal initiated NNCA polymerization in THF at room temperature. |

|
Download:
|
| Fig. 2. GPC traces of polypeptoids obtained in entries 6-10 in Table 1. | |
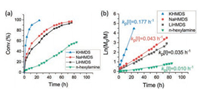
The polymerization rate difference was directly reflected in the study on NNCA conversion and reaction rate using inactive cyclohexyl-NNCA as the model (Fig. 3), which indicates that KHMDS has superior advantage as an initiator for inactive NNCA polymerization. The kp[I] value, as the reflection of reaction rate, for n-hexylamine-, LiHMDS-, NaHMDS- and KHMDS-initiated cyclohexyl-NNCA polymerization was 0.010 h−1, 0.035 h−1, 0.043 h−1 and 0.177 h−1, respectively, at a cyclohexyl-NNCA concentration of 1 mol/L and M/I ratio of 20. The substantially increased polymerization rate on cyclohexyl-NNCA using KHMDS as the initiator than n-hexylamine encourages us to further figure out the reaction mechanism.
|
Download:
|
| Fig. 3. Polymerization kinetics of cyclohexyl-NNCA polymerization initiated by n-hexylamine, LiHMDS, NaHMDS and KHMDS at room temperature, respectively. | |
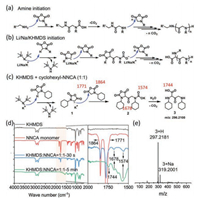
For the classical n-hexylamine strategy of NNCA polymerization, the reactive center for propagation is the secondary amine, which has low nucleophilicity to attack a new NNCA ring if the N-substitution group is bulky such as cyclohexyl, cyclopentyl and cyclooctyl (Fig. 4a). Therefore, the n-hexylamine-initiated polymerization on cyclohexyl-NNCA takes over 6 days as aforementioned. In sharp contrast, as we demonstrated in previous NCA polymerization to prepare polypeptides [36, 39], we proposed that Li/Na/KHMDS-initiated NNCA polymerization exploits terminal N-carbamate as the initiator, which is much less affected by the N-substitution group for nucleophilic attack to a new NNCA and, therefore, KHMDS led to the completion of cyclohexyl-NNCA within 20 h (Fig. 4b). Taking KHMDS for example, we proposed that KHMDS initiate the NNCA polymerization by deprotonating the CH on the NNCA ring, followed by nucleophilic attack of the generated carbanion to the carbonyl carbon on a new NNCA ring and a subsequent ring-opening of the new NNCA ring to give a terminal N-carbamate as the reactive center for further propagation (Fig. 4c). Such a polymerization initiation mechanism was supported by the in situ Fourier transform infrared (FT-IR) characterization on the 1:1 molar ratio mixture of cyclohexyl-NNCA and KHMDS, based on the disappearance of characteristic peaks for two carbonyl groups on the NNCA monomer ring at 1864 cm−1 and 1771 cm−1, and the appearance of peaks at 1678 cm−1 and 1574 cm−1 for compound 2, and the appearance of peak at 1744 cm−1 for compound 3. The HRESI-MS characterization also supported our proposed mechanism by observing the peaks for compound 3 at m/z of 297.2181 and 319.2001.
|
Download:
|
| Fig. 4. (a, b) NNCA polymerization initiated by primary amine and Li/Na/KHMDS, respectively. (c) Initiation study on cyclohexyl-NNCA polymerization using KHMDS as the initiator using a 1:1 molar ratio mixture of cyclohexyl-NNCA and KHMDS. (d, e) FT-IR and HRESI-MS characterization, respectively, on a 1:1 molar ratio mixture of cyclohexyl-NNCA and KHMDS. | |
When the obtained polypeptoids are subjected to purification, water in solvent could cause hydrolysis of C-terminal unreacted NNCA ring to give a C-terminal carboxylate, and the protonation and decarboxylation of N-terminal N-carbamate to give an N-terminal amine, which is confirmed by the MALDI-TOF mass characterization on the purified cyclohexyl-NNCA polypeptoid for the observation of high intensity peaks at m/z of 613.3, 752.5, 891.6 (Fig. 5a). Still we observed peaks of compounds a3, b3, c3, d3 from this MALDI-TOF mass characterization, which all supported our proposed mechanism of Li/Na/KHMDS-initiated NNCA polymerization. It is worth mentioning that we observed a peak at m/z of 734.5, could be the signal of cyclized polypeptoid f5, but at a very low intensity. The proposed Li/Na/KHMDS-initiated NNCA polymerization mechanism and the C-terminal unreacted NNCA ring imply that a C-terminal functionalization can be easily obtained to functionalize the C-termini of polypeptoids, which was supported by our proof-of-concept demonstration of C-terminal functionalization with tert-butyl-benzylamine in high efficiency, according to the proton integration at chemical shift of 7.24−7.35 ppm in proton NMR characterization (Fig. 5b). This almost quantitative C-terminal functionalization, with almost one C-terminal tert-butyl-benzylamine per polypeptoid chain, underpinned that this NNCA polymerization afford linear polypeptoids dominantly.
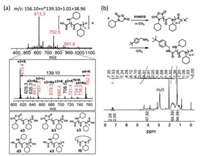
|
Download:
|
| Fig. 5. (a) MALDI-TOF mass characterization on polypeptoid that was obtained from KHMDS-initiated polymerization on cyclohexyl-NNCA. (b) In situ C-terminal functionalization of polypeptoid using KHMDS-initiated cyclohexyl-NNCA polymerization and resulting polypeptoid with almost quantitative C-terminal functionalization of tert-butyl-benzylamine. | |
In summary, polypeptoids have been explored as synthetic mimics of polypeptides for diverse functions and applications. Amines are widely used as initiators for NNCA polymerization to prepare polypeptoids. Nevertheless, new NNCA polymerization strategies are in great need, especially to address the slow polymerization on inactive NNCAs with bulky N-substitution group. Our discovery on alkali-metal initiated NNCA polymerization addresses this challenge with fast polymerization even on inactive NNCAs and can substantially enrich the structure diversity of polypeptoids for functional study and application.
Declaration of competing interestThe authors report no declarations of interest.
AcknowledgmentsThis research was supported by the National Natural Science Foundation of China (Nos. 22075078, 21774031, 21861162010), the Natural Science Foundation of Shanghai (No. 18ZR1410300), the National Key Research and Development Program of China (No. 2016YFC1100401), the National Natural Science Foundation of China for Innovative Research Groups (No. 51621002), Program of Shanghai Academic/Technology Research Leader (No. 20XD1421400), Research program of State Key Laboratory of Bioreactor Engineering, the Fundamental Research Funds for the Central Universities (No. 22221818014).
Appendix A. Supplementary dataSupplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.cclet.2021.02.039.
| [1] |
M. Xiong, Y. Bao, X. Xu, et al., Proc. Natl. Acad. Sci. U. S. A. 114 (2017) 12675-12680. DOI:10.1073/pnas.1710408114 |
| [2] |
L. Liu, K. Xu, H. Wang, et al., Nat. Nanotechnol. 4 (2009) 457-463. DOI:10.1038/nnano.2009.153 |
| [3] |
B.P. Mowery, S.E. Lee, D.A. Kissounko, et al., J. Am. Chem. Soc. 129 (2007) 15474-15476. DOI:10.1021/ja077288d |
| [4] |
S. Zhang, X.M. Xiao, F. Qi, et al., Chin. J. Polym. Sci. 37 (2019) 1105-1112. DOI:10.1007/s10118-019-2278-0 |
| [5] |
M. Zhou, X. Xiao, Z. Cong, et al., Angew. Chem. Int. Ed. 59 (2020) 7240-7244. DOI:10.1002/anie.202001697 |
| [6] |
T. Liu, Y.F. Zhang, S.Y. Liu, Chin. J. Polym. Sci. 31 (2013) 924-937. DOI:10.1007/s10118-013-1281-0 |
| [7] |
X. Wu, Y. Wu, H. Ye, et al., J. Control. Release 255 (2017) 81-93. DOI:10.1016/j.jconrel.2017.04.011 |
| [8] |
H. He, N. Zheng, Z.Y. Song, et al., ACS Nano 10 (2016) 1859-1870. DOI:10.1021/acsnano.5b05470 |
| [9] |
H. Xiang, Y. Chen, J. Zhang, et al., Chin. Chem. Lett. 31 (2020) 1422-1426. DOI:10.1016/j.cclet.2020.03.017 |
| [10] |
S. Han, Z. Li, J. Zhu, et al., Small 11 (2015) 2543-2554. DOI:10.1002/smll.201402865 |
| [11] |
J.G. Huang, J.C. Li, Y. Lyu, et al., Nat. Mater. 18 (2019) 1133-1143. DOI:10.1038/s41563-019-0378-4 |
| [12] |
Y. Ge, P. Li, Y. Guan, et al., Chin. Chem. Lett. 30 (2019) 1428-1431. DOI:10.1016/j.cclet.2019.03.009 |
| [13] |
Q. Chen, D. Zhang, W. Zhang, et al., Nat. Commun. 12 (2021) 562. DOI:10.1038/s41467-020-20858-x |
| [14] |
L. Wu, Y. Zou, C. Deng, et al., Biomaterials 34 (2013) 5262-5272. DOI:10.1016/j.biomaterials.2013.03.035 |
| [15] |
J.W. Checco, D.F. Kreitler, N.C. Thomas, et al., Proc. Natl. Acad. Sci. U. S. A. 112 (2015) 4552-4557. DOI:10.1073/pnas.1420380112 |
| [16] |
X. Zhou, Z. Li, Adv. Healthc. Mater. 7 (2018) 1800020. DOI:10.1002/adhm.201800020 |
| [17] |
B. Li, Y.M. Wu, W.J. Zhang, et al., Biomater. Sci. 7 (2019) 3675-3682. DOI:10.1039/C9BM00484J |
| [18] |
D. Zhang, Q. Chen, W. Zhang, et al., Angew. Chem. Int. Ed. 59 (2020) 9586-9593. DOI:10.1002/anie.202000416 |
| [19] |
T.H. Zhang, J.X. Yao, J.M. Tian, et al., Chin. Chem. Lett. 31 (2020) 1129-1132. DOI:10.1016/j.cclet.2019.07.010 |
| [20] |
Y. Liu, D. Li, J. Ding, et al., Chin. Chem. Lett. 31 (2020) 3001-3014. DOI:10.1016/j.cclet.2020.04.029 |
| [21] |
Z. Zheng, L. Zhang, Y. Ling, et al., Eur. Polym. J. 115 (2019) 244-250. DOI:10.1016/j.eurpolymj.2019.03.034 |
| [22] |
M. Zheng, M. Pan, W. Zhang, et al., Bioact. Mater. 6 (2021) 1878-1909. DOI:10.1016/j.bioactmat.2020.12.001 |
| [23] |
S.M. Miller, R.J. Simon, S. Ng, et al., Bioorg. Med. Chem. Lett. 4 (1994) 2657-2662. DOI:10.1016/S0960-894X(01)80691-0 |
| [24] |
J. Sun, R.N. Zuckermann, ACS Nano 7 (2013) 4715-4732. DOI:10.1021/nn4015714 |
| [25] |
X. Fu, C. Xing, J. Sun, Biomacromolecules 21 (2020) 4980-4988. DOI:10.1021/acs.biomac.0c01177 |
| [26] |
A.S. Knight, E.Y. Zhou, M.B. Francis, et al., Adv. Mater. 27 (2015) 5665-5691. DOI:10.1002/adma.201500275 |
| [27] |
C. Secker, S.M. Brosnan, R. Luxenhofer, et al., Macromol. Biosci. 15 (2015) 881-891. DOI:10.1002/mabi.201500023 |
| [28] |
R. Luxenhofer, C. Fetsch, A. Grossmann, Polym. Chem. 51 (2013) 2731-2752. DOI:10.1002/pola.26687 |
| [29] |
A. Li, L. Lu, X. Li, et al., Macromolecules 49 (2016) 1163-1171. DOI:10.1021/acs.macromol.5b02611 |
| [30] |
S. Varlas, P.G. Georgiou, P. Bilalis, et al., Biomacromolecules 19 (2018) 4453-4462. DOI:10.1021/acs.biomac.8b01326 |
| [31] |
L. Guo, S.H. Lahasky, K. Ghale, et al., J. Am. Chem. Soc. 134 (2012) 9163-9171. DOI:10.1021/ja210842b |
| [32] |
B.A. Chan, S. Xuan, A. Li, et al., Biopolymers 109 (2018) e23070. |
| [33] |
X. Fu, Z. Li, J. Wei, et al., Polym. Chem. 9 (2018) 4617-4624. DOI:10.1039/C8PY00924D |
| [34] |
D. Zhang, S.H. Lahasky, L. Guo, et al., Macromolecules 45 (2012) 5833-5841. DOI:10.1021/ma202319g |
| [35] |
J. Liu, J. Ling, J. Phys. Chem. A 119 (2015) 7070-7074. DOI:10.1021/acs.jpca.5b04654 |
| [36] |
Y. Wu, D. Zhang, P. Ma, et al., Nat. Commun. 9 (2018) 5297. DOI:10.1038/s41467-018-07711-y |
| [37] |
J.D. Ding, J. Funct. Polym. 32 (2019) 120-122. |
| [38] |
X.H. Wan, X.H. Wang, Acta Polym. Sinica 50 (2019) 99-101. DOI:10.11777/j.issn1000-3304.2018.18139 |
| [39] |
Y. Wu, W. Zhang, R. Zhou, et al., Chin. J. Polym. Sci. 38 (2020) 1131-1140. DOI:10.1007/s10118-020-2471-1 |
 2021, Vol. 32
2021, Vol. 32