b CAS Key Laboratory of Separation Science for Analytical Chemistry, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China
Mitochondria, which are generally considered as the power house of cell, are crucial and semi-autonomous organelles for energy production, cell respiration, regulation of metabolism and cell signaling [1, 2]. Therefore, real-time monitoring mitochondrial states can indicate the tiny changes of physiological process in living cells. Mitochondria can form dynamic network and are endowed with the ability to change their shape (elongation, shortening, branching, buckling, swelling) and location in three dimensions, which make it difficult to real-time monitor mitochondria in situ [3-5]. For instance, fusion and fission of mitochondria, closely related to stability of mtDNA and apoptosis, are dynamic events which usually occur in short time [6]. Besides, mitochondria can frequently communicate with other organelles, along with revelation of various physiological processes [7-9].
Although electron microscopy (EM), which boasts sub-nanometer resolution, has been widely used to inspect different form of mitochondria, the dynamic visualization of mitochondria in living cells is difficult to be achieved until the appearance of fluorescence microscopy. Conventionally, laser scan confocal microscopy has become a universal tool to monitor in situ events in living cells. However, the far-field optical diffraction limit restricts the spatial resolution beyond 200 nm [10-15]. Recently emerging super-resolution fluorescence microscopes, such as stimulated emission deletion (STED) [16], structured illumination microscopy (SIM) [17, 18], and stochastic optical reconstruction microscopy (STORM) [19], can break the conventional resolution limit which allow the visualization of dynamic mitochondria at the nanoscale in cells. Among them, SIM is a widefield microscopy technique with a spatial resolution of up to 100 nm which is realized by illuminating with multiple interfering beams of light. Furthermore, SIM has gradually become one of key technologies in modern cell biology, especially in dynamic changes of mitochondria, for the preferable biocompatibility and no extra demands in experimental procedures. But the ideal long-term dynamic super-resolution imaging of mitochondria requires excellent accuracy and stability of labeling [20], but remains challenging. Nowadays, organic probes with higher brightness and photostability compared with fluorescent proteins are promisingly used to label mitochondria in SIM imaging. Zhang's group used an existing cell-permeable organic fluorescent probes, Atto 647 N dye, to achieve specific labeling of mitochondria in live cells [21]. Through dual-color SIM images of dynamic physical lysosome-mitochondrion interactions, the authors observed consecutive dynamic processes of lysosomal fusion and fission. In addition, Diao and coworkers proposed a strategy to screen and discover drugs by monitoring of mitochondria and lysosomes interactions at nanoscale level in living cells using Mito-Tracker Green [22]. However, most mitochondria-targeted probes are lipophilic cations with so called covalent linkage sites which are usually considered as mitochondrial membrane potential probes [23-25]. The accuracy and stability of mitochondria labeling mainly depend on the negative mitochondrial membrane potential. As a result, once the mitochondrial membrane potential changed, redundant probes would dissociate to cytoplasm which will greatly increase the background. In another words, existing mitochondrial membrane potential probes can hardly achieve long term and dynamic superresolution imaging of mitochondria, especially in mitophagy, apoptosis which can induce loss of membrane potential.
In our previous research, we reported a series of wash-free SNAP-tag fluorogenic probe based on naphthalimide which could specifically label mitochondria through the formation of stable thioether bond between probes and SNAP-Cox8A fusion proteins [26, 27]. For this reason, the probes could realize long term and dynamic imaging of mitochondria without diffusion in living cells. Herein, we developed a novel SNAP-tag fluorogenic probe, which could achieve enormous fluorescence enhancement upon labeling SNAP-tag (Scheme 1), based on 4-azetidinyl-naphthalimide derivatives. Through the stable fluorescence labeling of mitochondria and high signal-to-noise ratio, dynamic mitochondria were visualized by SIM in living cells without washing. Furthermore, fusion and fission of mitochondria were also monitored at nanoscale level. We also monitored the fusion of mitochondria and lysosomes during mitophagy via dual-color SIM imaging with the wash-free SNAP-tag probe. This strategy to visualize dynamic mitochondria at nanoscale level will provide deep understanding of mitochondrial function in physiological process. The experimental procedures can be found in Supporting information.

|
Download:
|
| Scheme 1. Schematic illustration of SNAP-tag sensing. | |
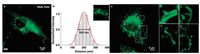
We first investigated whether the probe could realize wash-free SIM imaging of SNAP-tag fusion proteins in mitochondria. Commercial available plasmids pSNAPf-Cox8A fusing SNAP-tag to cytochrome c oxidase subunit 8 (Cox8A) was transiently transfected to HeLa cells (the cell culture and transfection methods can be found in Supporting information). After incubated with the SNAP-tag probe for 1 h, HeLa cells were directly imaged without washing processes. Strong green fluorescence was observed in different form of mitochondria, while other locations and non-transfected cells showed negligible background under wide-field imaging (Fig. 1a). The SIM images displayed a greatly improved spatial resolution (~109 nm, Fig. 1b) in which mitochondrial cristae were clearly distinguished (Fig. 1c, 1–4). Furthermore, the network of mitochondria, consisted with rounded, clavate and linear mitochondria, was extraordinary coherent. It was revealed that the probe was able to image mitochondria using SIM in living cells without washing.
|
Download:
|
| Fig. 1. (a) Comparison of SIM image and wild field image of the cell in Fig. 2. (b) Cross-sectional profile of a mitochondrial cristae at 16 min in Fig. 2 at a full-width at the halfmaximum (FWHM) of 109 nm. Excitation: 488 nm, collected: 500-545 nm. (c) Wash-free SIM images of living Hela cells incubated with 1 μmol/L SNAP-tag fluorogenic probe. (1), (2), (3) and (4) Enlargements of the boxed regions in (c), in which the fine structures especially mitochondrial cristae were visible. | |
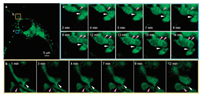
The excellent performance in SIM imaging inspired us to continuously monitor dynamic changes of mitochondria in living cells. During the time course of over 16 min, the green fluorescence from mitochondria was almost unchanged and the outline of mitochondria were clearly demarcated (Fig. 2). Besides, we observed the shape changes of mitochondria which varied from linear to irregular mitochondria, and to rounded mitochondria finally (Fig. 2b, pink arrow). Moreover, the process of mitochondrial fusion and fission was long term monitored in living cells. As shown in Fig. 2b, as time went on, the site labeled by white arrow changed to slender structure and reached to fracture at the 12th min. So, the single mitochondrion, which exhibited branch at the 1 st min, gradually turned into two dumbbell-like mitochondria. Furthermore, the fusion of two isolated mitochondria was viewed in the same cell (Fig. 2c). In the first 5 min, the two mitochondria were found to establish communications through the slender structure labeled by white arrow, which became coarse next. Two obvious mitochondrial cristae were gradually formed along with the mitochondrial fusion, due to the accumulation of matrix compartment during fusion of two mitochondria material of the inner membrane. Through SNAP-tag fluorogenic probe, long-term super-resolution imaging of dynamic mitochondria was realized which might provide functional significance of mitochondria in mtDNA mutations and programmed cell death.
|
Download:
|
| Fig. 2. Time-lapse SIM images of living HeLa cells stained with SNAP-tag probe. (a) The SIM image of whole living HeLa cells. (b, c) Enlargements of the boxed regions in (a), which revealed different types of dynamic physical process of mitochondria. (b) SIM images of mitochondrial morphologic changes of mitochondria swollen with laser irradiation. (c) SIM images of mitochondria fission (white arrow) and fusion (pink arrow). For the time-lapse images, the time interval between each SIM image was set to 30 s. Excitation: 488 nm, collected: 500-545 nm. | |
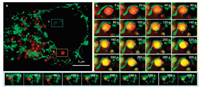
Finally, we intended to monitor the fusion of mitochondria and lysosomes during mitophagy which could maintain homeostasis through selectively degrading damaged mitochondria. Therefore, long-term dual-color SIM imaging of mitochondria and lysosomes in Hela cells was performed (Fig. 3). In order to facilitate the occurrence of mitophagy, we enhanced the power of laser (0.5 kW/cm2) for laser-induce damage was reported as one of effective method stoinduce mitophagy.Initially, thelysosomelabeled by Lyso-Tracker Red and mitochondria were completely separated (Fig. 3b, 0 s). However, ~40 s later, the mitochondria-lysosome contact increases, as evidenced by the nearness between mitochondria and lysosomes. Hence, the locally damaged mitochondria could be transported to lysosome which indicated the accomplishment of mitophagy along with the increase of yellow area in circle. In the meanwhile, numbers of mitophagy were captured at nanoscale level in living cells (Fig. 3c).
|
Download:
|
| Fig. 3. Dual-color SIM images of lysosomes and mitochondria in living HeLa cells. (a) The SIM image of whole live Hela cells stained with Lyso-Tracker Red (red) and SNAP-tag fluorogenic probe (green). (b) and (c) are representative time-lapse SIM images of different types of dynamic physical interactions between lysosomes and mitochondria in live Hela cells. For the time-lapse images, the time interval between each SIM image was set to 20 s. Excitation: 488 nm, collected: 500-545 nm (green channel). Excitation: 561 nm, collected: 570-640 nm (red channel). | |
In conclusion, we introduced a new method for long term monitoring dynamic mitochondria via SNAP-tag at nanoscale level. The SNAP-tag fluorogenic probe could realize wash-free SIM imaging of mitochondria in living cells without diffusion due to the formation of stable thioether bond with SNAP-Cox8A fusion proteins. Furthermore, we recorded consecutive dynamic physical process of mitochondrial fusion and fission with our SNAP-tag probe over a course of 16 min. The formation of mitochondrial cristae, due to the accumulation of matrix compartment, during fusion was also clearly demonstrated. Meanwhile, we successfully applied this method to track the interaction of mitochondria and lysosomes, and captured the fusion between mitochondria and lysosomes during mitophagy in live cells. The method in this paper for nanoscale monitoring dynamic mitochondria in living cells offers a convincing mode to indicate the role of mitochondria in molecular biological mechanisms.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentsThis work was supported by the National Natural Science Foundation of China (Nos. 21878286, 21576043, 21878286), Dalian Institute of Chemical Physics, Chinese Academy of Sciences (Nos. I201938, ZZBS201805).
Appendix A. Supplementary dataSupplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.cclet.2020.05.043.
| [1] |
Bereiter-Hahn J., M. Vöth, Microsc. Res. Tech. 27 (1994) 198-219. DOI:10.1002/jemt.1070270303 |
| [2] |
B. Westermann, W. Neupert, Yeast 16 (2000) 1421-1427. DOI:10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U |
| [3] |
H.M. McBride, M. Neuspiel, S. Wasiak, Curr. Biol. 16 (2006) R551-R560. DOI:10.1016/j.cub.2006.06.054 |
| [4] |
L.C. Gomes, G.D. Benedetto, L. Scorrano, Nat. Cell Biol. 13 (2011) 589-598. DOI:10.1038/ncb2220 |
| [5] |
S. Hoppins, L. Lackner, J. Nunnari, Annu. Rev. Biochem. 76 (2007) 751-780. DOI:10.1146/annurev.biochem.76.071905.090048 |
| [6] |
D.C. Chan, Cell 125 (2006) 1241-1252. DOI:10.1016/j.cell.2006.06.010 |
| [7] |
I.R. Boldogh, L.A. Pon, Trends Cell Biol. 17 (2007) 502-510. DOI:10.1016/j.tcb.2007.07.008 |
| [8] |
M. Audano, A. Schneider, N. Mitro, J. Neurochem. 147 (2018) 291-309. DOI:10.1111/jnc.14471 |
| [9] |
Y.C. Wong, S. Kim, W. Peng, D. Krainc, Trends Cell Biol. 29 (2019) 500-513. DOI:10.1016/j.tcb.2019.02.004 |
| [10] |
S.W. Hell, Science 316 (2007) 1153-1158. DOI:10.1126/science.1137395 |
| [11] |
B. Huang, H. Babcock, X. Zhuang, Cell 143 (2010) 1047-1058. DOI:10.1016/j.cell.2010.12.002 |
| [12] |
Q. Hu, C. Qin, L. Huang, H. Wang, Q. Liu, L. Zeng, et al., Dyes Pigments 149 (2018) 253-260. DOI:10.1016/j.dyepig.2017.10.002 |
| [13] |
J. Xu, J. Pan, X. Jiang, et al., Biosens. Bioelectron. 77 (2016) 725-732. DOI:10.1016/j.bios.2015.10.049 |
| [14] |
F. Deng, Z. Xu, Chin. Chem. Lett. 30 (2019) 1667-1681. DOI:10.1016/j.cclet.2018.12.012 |
| [15] |
S. Long, Q. Qiao, L. Miao, Z. Xu, Chin. Chem. Lett. 30 (2019) 573-576. DOI:10.1016/j.cclet.2018.11.031 |
| [16] |
F. Huang, T.M.P. Hartwich, Rivera-Molina F.E., et al., Nat. Methods 10 (2013) 653-658. DOI:10.1038/nmeth.2488 |
| [17] |
L. Schermelleh, P.M. Carlton, S. Haase, et al., Science 320 (2008) 1332-1336. DOI:10.1126/science.1156947 |
| [18] |
X. Huang, J. Fan, L. Li, et al., Nat. Biotechnol. 36 (2018) 451-459. DOI:10.1038/nbt.4115 |
| [19] |
M.J. Rust, M. Bates, X. Zhuang, Nat. Methods 3 (2006) 793-796. DOI:10.1038/nmeth929 |
| [20] |
H. Takakura, Y. Zhang, R.S. Erdmann, et al., Nat. Biotechnol. 35 (2017) 773-780. DOI:10.1038/nbt.3876 |
| [21] |
Y. Han, M. Li, F. Qiu, M. Zhang, Y.H. Zhang, Nat. Commun. 8 (2017) 1307. DOI:10.1038/s41467-017-01503-6 |
| [22] |
Q. Chen, X. Shao, Z. Tian, et al., Nano Res. 12 (2019) 1009-1015. DOI:10.1007/s12274-019-2331-x |
| [23] |
R.A.J. Smith, R.C. Hartley, M.P. Murphy, Antioxid. Redox Signal. 15 (2011) 3021-3038. DOI:10.1089/ars.2011.3969 |
| [24] |
J. Zielonka, J. Joseph, A. Sikora, et al., Chem. Rev. 117 (2017) 10043-10120. DOI:10.1021/acs.chemrev.7b00042 |
| [25] |
S. Long, W. Chi, L. Miao, et al., Chin. Chem. Lett. 30 (2019) 601-604. DOI:10.1016/j.cclet.2018.12.008 |
| [26] |
S. Leng, Q. Qiao, L. Miao, et al., Chem. Commun. 53 (2017) 6448-6451. DOI:10.1039/C7CC01483J |
| [27] |
Q. Qiao, W. Liu, J. Chen, et al., Dyes Pigments 147 (2017) 327-333. DOI:10.1016/j.dyepig.2017.08.032 |
 2020, Vol. 31
2020, Vol. 31 

