b Guangdong Provincial Key Laboratory of Laser Life Science, College of Biophotonics, South China Normal University, Guangzhou 510631, China;
c Zhuhai Interventional Medical Center, Zhuhai Precision Medical Center, Zhuhai People's Hospital, Zhuhai Hospital Affiliated with Jinan University, Jinan University, Zhuhai 519000, China
Pathogenic bacteria can cause serious morbidity and have become a critical public healthcare problem [1, 2]. Especially, the infections caused by multidrug-resistant bacteria, are a global burden resulting in huge human death in the hundreds of thousands every year [3, 4]. The emergence of resistant and more virulent bacteria strains even surpasses the development of new antimicrobial agents and methods [5]. Therefore, it is extremely urgent to develop new materials and alternative treatment strategiesto combatantibiotic resistance [6-9]. Although photodynamic therapy (PDT) [10-12], sonodynamic therapy (SDT) [13] and photothermal therapy (PTT) [14, 15] are promising methods to solve the issue of bacterial resistance, each of these is frequently suffered from some typical shortcomings, such as low efficiencies, low laser penetration depth, and poor therapeutic efficacy of single treatmentmodality [16].Hence, it is still challenging to develop efficient therapeutic methods for bacterial killing. Multimodal therapy, the combination of two or more different therapeutic modalities, has become a promising approach to enhance antibacterial efficiency [17, 18].
Typically, antimicrobial photodynamic therapy, as a photoinduced therapy, has been successfully applied in epidermis microbial infection [19-21]. Antimicrobial photodynamic therapy possesses some advantages, including non-invasiveness, easy operation, short treatment cycle, low side effects and overcoming of resistant bacteria [22-24]. Resistant bacteria can be easily destroyed due to indiscriminate photodynamic damage to bacterial components [25, 26], which results from the generated reactive oxygen species (ROS) that are toxic toward cells and microbes [27]. Most photosensitizers show good antibacterial ability against Gram-positive bacteria, but they are insufficient to inhibit Gram-negative bacteria because of their distinct membrane structure [28, 29]. Therefore, it is imperative to develop broad-spectrum antibacterial materials that can inhibit both Gram-positive and Gram-negative bacteria [30].
Polyhexamethylene guanidine (PHMG), an effective and broad-spectrum cationic polymeric bactericide, has been widely applied in many fields (Scheme 1a), including medicine [31, 32], wound care [33], water treatment [34], food industry, paper making and so on [35]. The antimicrobial mechanism of PHMG is demonstrated that cationic polymers can rapidly adsorb onto the negatively charged cell surface, impair the cellular membrane, and increase the permeability of cytoplasmic membrane to in-situ result in the formation of local pores, the leakage of intracellular components and final bacterial death [36-39]. Importantly, the physical damage mechanism of PHMG is very different from traditional antibiotics [40, 41]. Physical damage is promising to overcome the bacterial resistance [42-44]. Meantime, the terminal amino groups of PHMG provide reactive sites to conjugate with other functional units.
|
Download:
|
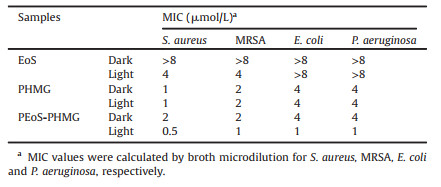
| Scheme 1. Schematic illustration for (a) the synthesis of Eosin-Y polymer conjugate-modified PHMG, P(DMAEMA-co-EoS)-b-PHMG-b-P(DMAEMA-co-EoS), noted as PEoS-PHMG. (b) Dual-modality inhibition broad-spectrum bacteria via synergistic photodynamic inactivation and physical damage. | |
In this work, we reported an efficient polymeric antimicrobial of EoS photosensitizer conjugate-functionalized PHMG, P(DMAEMA-co-EoS)-b-PHMG-b-P(DMAEMA-co-EoS), noted as PEoS-PHMG, which was prepared from the amidation reaction of PHMG and P(DMAEMA-co-EoS). Owing to the conjugation with P(DMAEMA-co-EoS), the resulting PEoS-PHMG exhibited dualmodality inhibition, including physical damage to the microbes and concurrent photodynamic killing (Scheme 1). PEoS-PHMG behaved great antimicrobial activities against methicillin-resistant Staphylococcus aureus (MRSA) and typical Gram-negative bacteria. Additionally, the antimicrobial PEoS-PHMG had good biocompatibility and low toxicity to mammalian cells.
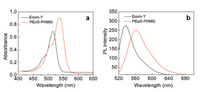
The fabrication of PEoS-PHMG was illustrated in Scheme 1a. The EoS-labeled polymer conjugate, P(DMAEMA-co-EoS) was prepared by facile reversible addition fragmentation chain transfer (RAFT) polymerization at first [45, 46], which was characterized by 1H NMR spectrum (Fig. S1 in Supporting information). The covalent conjugation between P(DMAEMA-co-EoS) and PHMG was achieved by the amidation reaction, affording PEoS-PHMG. The successful modification was characterized by typical FT-IR analysis (Fig. S2 in Supporting information), indicating the stretching vibration signals of C=N and C=O bonds in the resultant PEoSPHMG. In addition, the UV-vis absorption spectra of Eosin-Y and PEoS-PHMG were recorded in water (Fig. 1a), indicating the absorbance red-shift from ~516 nm to ~536 nm for EoS moieties in PEoS-PHMG, which also confirmed the successful conjugation of P(DMAEMA-co-EoS) with PHMG. For the fluorescence emission spectra (Fig. 1b), similar emission red-shift from ~535 nm to ~562 nm for EoS moieties was also observed. In addition, the mean hydrodynamic diameter for the aqueous dispersion of PEoS-PHMG was determined to be ~12 nm by dynamic light scattering (DLS) analysis (Fig. S3 in Supporting information).
|
Download:
|
| Fig. 1. (a) Absorbance spectra and (b) fluorescence emission spectra recorded for Eosin-Yand PEoS-PHMG, respectively. The excitation wavelength was set at 500 nm. | |
To verify the photodynamic potency of PEoS-PHMG, the single oxygen (1O2) generation potency was performed using 1, 3-dipheny-lisobenzofuran (DPBF) as an indicator of 1O2 [47, 48]. DPBF can react with 1O2 in a stoichiometric manner and form an irreversible endoperoxide to cause absorption decrease at ~410 nm [9]. As shown in Figs. 2a-c, after exposure to light, obvious absorption decreasing at ~410 nm was observed for Eosin-Y and PEoS-PHMG. More importantly, the consumption of DPBF for the PEoS-PHMG group was more quickly than that of Eosin-Y, suggesting enhanced 1O2 yield for the resultant PEoS-PHMG. In contrast, no obvious absorbance decrease of DPBF was determined for PHMG. The normalized absorbance variation of different groups was also shown in Fig. 2d. These results indicated that PEoS-PHMG had the best ROS generation efficiency.

|
Download:
|
| Fig. 2. In vitro evaluation of ROS generation upon light irradiation (520 ±10 nm) at diverse durations for (a) PHMG, (b) Eosin-Y, and (c) PEoS-PHMG, respectively. DPBF was employed as a ROS indicator for ROS evaluation. (d) Normalized absorbance intensity at 410 nm recorded for (a)-(c), respectively. | |
The antimicrobial activities of these materials were evaluated against clinically representative bacteria, including Gram-positive bacteria, S. aureus, MRSA and Gram-negative bacteria, E. coli and P. aeruginosa. Using a broth microdilution protocol [49], the minimum inhibitory concentrations (MIC) values were presented in Table 1 and Fig. 3, Figs. S5-S7 (Supporting information). These results demonstrated that PEoS-PHMG exhibited broad-spectrum antimicrobial activities against S. aureus, MRSA, E. coli and P. aeruginosa with MICs values of 0.5, 1, 1, 1 μmol/L under light irradiation, respectively. The MIC values of PEoS-PHMG were much lower than PHMG (1, 2, 4, 4 μmol/L) owing to that PHMG did not possess photodynamic antibacterial potency, only exhibiting physical inhibition for PHMG. Upon green light irradiation to activate the photodynamic process, the inhibition ability of Eosin-Y was still limited, especially for Gram-negative bacteria, because the generated ROS could not approach and attack bacteria membrane efficiently due to the short life time and limited action range of ROS [50, 51]. Furthermore, Gram-negative bacteria possess complicated cell envelopes with low permeability and extra defensive mechanisms [52, 53]. Hence, the dual-modality antimicrobial potency of PEoS-PHMG exhibited highly effective broad-spectrum antibacterial ability.
|
|
Table 1 Minimum inhibition concentration (MIC) values evaluated for EoS, PHMG and PEoS-PHMG, respectively. |

|
Download:
|
| Fig. 3. Quantitative antibacterial tests were performed against E. coli upon incubation with different contents of Eosin-Y, PHMG, and PEoS-PHMG under dark (a) and light irradiation (b), respectively. | |
To evaluate the synergistic bacterial inhibition of PEoS-PHMG via physical damage and photodynamic inactivation, typical combination index (CI) was utilized as a parameter via the Chou-Talalay method [54]. The CI value reflects the interaction effect of the dual-modality inhibition mechanisms. 0.3 < CI < 0.7 indicates obvious synergism, 0.7 < CI < 0.85 indicates moderate synergism [55]. The CI values of PEoS-PHMG with light irradiation for S. aureus, MRSA, E. coli, P. aeruginosa were calculated to be 0.375, 0.75, < 0.375, and < 0.375, respectively, indicating obvious synergy for S. aurues, E.coli, P.aeruginosa, and moderate effect for MRSA. These results demonstrated effective synergy for PEoS-PHMG via physical damage and PDT to inhibit broad-spectrum bacteria.
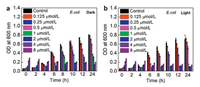
After that, to further verify the antibacterial effect of PEoS-PHMG, the OD600 values were recorded for the dispersion at different durations (Fig. 4). At the corresponding MIC value, there was no obvious OD600 rise in the whole process for the dispersion of E. coli upon incubation with PEoS-PHMG under light irradiation, exhibiting efficient inhibition. These results confirmed that the functionalization of PHMG further potentiated its broad-spectrum bacterial inhibition.
|
Download:
|
| Fig. 4. Time-dependent growth inhibition of E. coli after incubation with PEoS-PHMG (a) at dark and (b) light irradiation, respectively. | |
Furthermore, inhibition zone assay was performed in parallel for Eosin-Y, PHMG, and PEoS-PHMG, respectively (Fig. S4 in Supporting information). The area of inhibition zone reflected their relative inhibition potency. PEoS-PHMG possessed the largest inhibition zone upon light irradiation, suggesting favorable synergistical bacteria inhibition by PEoS-PHMG, which was much better than PHMG and Eosin-Y.
Apart from bacterial inhibition, the safety issue of antibacterial materials is critically important [56]. Then the hemolysis was analyzed for P(DMAEMA-co-EoS), PHMG and PEoS-PHMG, respectively. HC10, the sample content that causes 10% hemolysis of mouse red blood cells (mRBCs) relative to that of control, was determined by incubation mRBCs with different contentof the samples [57]. The HC10 value of PEoS-PHMG was determined to be over 32 μmol/L, and that of PHMG was detected to be higher than 16 μmol/L (Fig. 5). Hence, the blood safety of PEoS-PHMG was further enhanced for PHMG after modification [58, 59]. The presence of hydrophilic P(DMAEMA-co-EoS) with less positive charge probably contributed to the minimal hemolysis of PEoS-PHMG [60, 61].

|
Download:
|
| Fig. 5. Hemolysis activity analyzed for P(DMAEMA-co-EoS), PHMG and PEoS-PHMG, respectively. | |
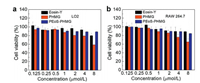
In vitro cell viability was tested for two kinds of cell lines after incubation with these diverse samples. Cell counting kit-8 (CCK-8) assays were performed for two series of samples upon incubation with LO2 cells and RAW 264.7 cells for 24 h (Fig. 6). PEoS-PHMG showed undetectable toxicity toward both LO2 and RAW 264.7 cells even up to 8 μmol/L, approximately 2~4-fold compared to their corresponding MIC values. The results demonstrated that the functionalized PEoS-PHMG had good antibacterial selectivity, which was toxic to bacteria but minimally harmful to mammalian cells.
|
Download:
|
| Fig. 6. In vitro cell viability of (a) LO2 cells and (b) RAW 264.7 cells after 24 h incubation with Eosin-Y, PHMG, and PEoS-PHMG at various contents. | |
In summary, a broad-spectrum polymeric antimicrobial, PEoS-PHMG, was developed to achieve dual-modality bacterial inhibition via photodynamic inactivation and physical damage. Compared with native PHMG, current photosensitizer conjugate-functionalized PHMG exhibited excellent bacterial inhibition and enhanced biocompatibility with minimal cytotoxicity toward mammalian cells. Herein, the photodynamic functionalization of PHMG is a promising strategy to exploit novel polymeric antimicrobials in biomedicine.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentsThis work was supported by the Natural Science Foundation for Distinguished Young Scholars of Guangdong Province (No. 2016A030306013), the Pearl River Young Talents Program of Science and Technology in Guangzhou (No. 201906010047) and the National Key Research and Development Program of China (No. 2017YFA0205200)
Appendix A. Supplementary dataSupplementary material related to this article can befound, in the online version, at doi:https://doi.org/10.1016/j.cclet.2020.06.038.
| [1] |
J. Zhou, G.B. Qi, H. Wang, J. Mater. Chem. B:Mater. Biol. Med. 4 (2016) 4855-4861. DOI:10.1039/C6TB00406G |
| [2] |
R. Yin, T. Agrawal, U. Khan, et al., Nanomedicine 10 (2015) 2379-2404. DOI:10.2217/nnm.15.67 |
| [3] |
A.S. Abd-El-Aziz, C. Agatemor, N. Etkin, et al., Biomacromolecules 16 (2015) 3694-3703. DOI:10.1021/acs.biomac.5b01207 |
| [4] |
L.J. Shallcross, S.J. Howard, T. Fowler, S.C. Davies, Philos. Trans. R. Soc. B 370 (2015) 20140082. DOI:10.1098/rstb.2014.0082 |
| [5] |
A. Reinhardt, I. Neundorf, Int. J. Mol. Sci. 17 (2016) 701. DOI:10.3390/ijms17050701 |
| [6] |
K.H. Wu, Y.C. Chang, J.C. Wang, Mater. Express 9 (2019) 970-977. DOI:10.1166/mex.2019.1569 |
| [7] |
A. Sun, X.Y. He, L. Li, et al., NPG Asia Mater. 12 (2020) 25. DOI:10.1038/s41427-020-0206-y |
| [8] |
X.F. Luo, Y.J. Liu, Z.D. Qin, et al., J. Biomed. Nanotechnol. 14 (2018) 601-608. DOI:10.1166/jbn.2018.2528 |
| [9] |
F. Xu, M. Hu, C. Liu, S.K. Choi, Biomater. Sci. 5 (2017) 678-685. DOI:10.1039/C7BM00030H |
| [10] |
X.J. Fu, Y. Fang, M. Yao, Biomed Res. Int. 2013 (2013) 159157. |
| [11] |
D. Hu, Y. Deng, F. Jia, Q. Jin, J. Ji, ACS Nano 14 (2020) 347-359. DOI:10.1021/acsnano.9b05493 |
| [12] |
K. Liu, Y. Liu, Y. Yao, et al., Angew. Chem. Int. Ed. 52 (2013) 8285-8289. DOI:10.1002/anie.201303387 |
| [13] |
D. Costley, H. Nesbitt, N. Ternan, et al., Int. J. Antimicrob. Agents 49 (2017) 31-36. DOI:10.1016/j.ijantimicag.2016.09.034 |
| [14] |
X. Huang, G. Chen, J. Pan, et al., J. Mater. Chem. B:Mater. Biol. Med. 4 (2016) 6258-6270. DOI:10.1039/C6TB01122E |
| [15] |
X. Guo, B. Cao, C. Wang, S. Lu, X. Hu, Nanoscale 12 (2020) 7651-7659. DOI:10.1039/D0NR00181C |
| [16] |
Y. Liu, H.C. van der Mei, B. Zhao, et al., Adv. Funct. Mater. 27 (2017) 1701974. DOI:10.1002/adfm.201701974 |
| [17] |
M. Yin, Z. Li, E. Ju, et al., Chem. Commun. (Camb.) 50 (2014) 10488-10490. DOI:10.1039/C4CC04584J |
| [18] |
F.E. Akram, T. El-Tayeb, K. Abou-Aisha, M. El-Azizi, Ann. Clin. Microbiol. Antimicrob. 15 (2016) 48. DOI:10.1186/s12941-016-0164-y |
| [19] |
M. van Oosten, M. Hahn, L.M. Crane, et al., FEMS Microbiol. Rev. 39 (2015) 892-916. DOI:10.1093/femsre/fuv029 |
| [20] |
G. Fila, K. Kasimova, Y. Arenas, et al., Front. Microbiol. 7 (2016) 1258. |
| [21] |
J. Xia, W. Wang, X. Hai, et al., Chin. Chem. Lett. 30 (2019) 421-424. DOI:10.1016/j.cclet.2018.07.008 |
| [22] |
L. Yang, P. Gao, Y. Huang, et al., Chin. Chem. Lett. 30 (2019) 1293-1296. DOI:10.1016/j.cclet.2019.03.032 |
| [23] |
T. Maisch, Lasers Med. Sci. 22 (2007) 83-91. DOI:10.1007/s10103-006-0409-7 |
| [24] |
X. Liu, M. Li, T. Han, et al., J. Am. Chem. Soc. 141 (2019) 11259-11268. DOI:10.1021/jacs.9b04757 |
| [25] |
H. Bai, F. Lv, L. Liu, S. Wang, Chemistry 22 (2016) 11114-11121. DOI:10.1002/chem.201600877 |
| [26] |
F. Vatansever, W.C. de Melo, P. Avci, et al., FEMS Microbiol. Rev. 37 (2013) 955-989. DOI:10.1111/1574-6976.12026 |
| [27] |
L. Chen, H. Bai, J.F. Xu, S. Wang, X. Zhang, ACS Appl. Mater. Interfaces 9 (2017) 13950-13957. DOI:10.1021/acsami.7b02611 |
| [28] |
X. Ragas, D. Sanchez-Garcia, R. Ruiz-Gonzalez, et al., J. Med. Chem. 53 (2010) 7796-7803. DOI:10.1021/jm1009555 |
| [29] |
J. Chen, F. Wang, Q. Liu, J. Du, Chem. Commun. (Camb.) 50 (2014) 14482-14493. DOI:10.1039/C4CC03001J |
| [30] |
X.K. Ding, S. Duan, X.J. Ding, R.H. Liu, F.J. Xu, Adv. Funct. Mater. 28 (2018) 1802140. DOI:10.1002/adfm.201802140 |
| [31] |
C. Kratzer, S. Tobudic, W. Graninger, A. Buxbaum, A. Georgopoulos, J. Hosp. Infect. 63 (2006) 316-322. DOI:10.1016/j.jhin.2006.01.024 |
| [32] |
M. Rosin, A. Welk, O. Bernhardt, et al., J. Clin. Periodontol. 28 (2001) 1121-1126. DOI:10.1034/j.1600-051X.2001.281206.x |
| [33] |
M.K. Oule, R. Azinwi, A.M. Bernier, et al., J. Med. Microbiol. 57 (2008) 1523-1528. DOI:10.1099/jmm.0.2008/003350-0 |
| [34] |
J.M. Kusnetsov, A.I. Tulkki, H.E. Ahonen, P.J. Martikainen, J. Appl. Microbiol. 82 (1997) 763-768. DOI:10.1046/j.1365-2672.1997.00151.x |
| [35] |
Y. Guan, H. Xiao, H. Sullivan, A. Zheng, Carbohydr. Polym. 69 (2007) 688-696. DOI:10.1016/j.carbpol.2007.02.013 |
| [36] |
P. Gilbert, L.E. Moore, J. Appl. Microbiol. 99 (2005) 703-715. DOI:10.1111/j.1365-2672.2005.02664.x |
| [37] |
Y. Wu, J. Bai, K. Zhong, Y. Huang, H. Gao, Food Chem. 218 (2017) 463-470. DOI:10.1016/j.foodchem.2016.07.090 |
| [38] |
Z. Liang, M. Zhu, Y.W. Yang, H. Gao, Polym. Advan. Technol. 25 (2014) 117-122. DOI:10.1002/pat.3212 |
| [39] |
Q. Cai, S. Yang, C. Zhang, et al., ACS Appl. Mater. Interfaces 10 (2018) 38506-38516. DOI:10.1021/acsami.8b14986 |
| [40] |
G.B. Qi, D. Zhang, F.H. Liu, Z.Y. Qiao, H. Wang, Adv. Mater. 29 (2017) 1703461. DOI:10.1002/adma.201703461 |
| [41] |
Q. Zeng, Y. Zhu, B. Yu, et al., Biomacromolecules 19 (2018) 2805-2811. DOI:10.1021/acs.biomac.8b00399 |
| [42] |
A. Azizi, S. Mousavian, S. Taheri, et al., Photodiagn. Photodyn. Ther. 21 (2018) 357-362. DOI:10.1016/j.pdpdt.2018.01.014 |
| [43] |
Y.Y. Huang, A. Wintner, P.C. Seed, et al., Sci. Rep. 8 (2018) 7257. DOI:10.1038/s41598-018-25365-0 |
| [44] |
H. Sun, Y. Hong, Y. Xi, et al., Biomacromolecules 19 (2018) 1701-1720. DOI:10.1021/acs.biomac.8b00208 |
| [45] |
K.L. Zhu, G.H. Liu, J.M. Hu, S.Y. Liu, Biomacromolecules 18 (2017) 2571-2582. DOI:10.1021/acs.biomac.7b00693 |
| [46] |
G.H. Liu, J.M. Hu, G.Q. Zhang, S.Y. Liu, Bioconjug. Chem. 26 (2015) 1328-1338. DOI:10.1021/bc500548r |
| [47] |
M. Yin, Z. Li, L. Zhou, et al., Nanotechnology 27 (2016) 125601. DOI:10.1088/0957-4484/27/12/125601 |
| [48] |
A. Gomes, E. Fernandes, J.L. Lima, J. Biochem, Biophys. Methods. 65 (2005) 45-80. DOI:10.1016/j.jbbm.2005.10.003 |
| [49] |
I. Wiegand, K. Hilpert, R.E.W. Hancock, Nat. Protoc. 3 (2008) 163-175. DOI:10.1038/nprot.2007.521 |
| [50] |
S. Perni, P. Prokopovich, J. Pratten, I.P. Parkin, M. Wilson, Photochem. Photobiol. Sci. 10 (2011) 712-720. DOI:10.1039/c0pp00360c |
| [51] |
Y. Guo, S. Rogelj, P. Zhang, Nanotechnology 21 (2010) 065102. DOI:10.1088/0957-4484/21/6/065102 |
| [52] |
Y.M. Li, X.L. Hu, S.D. Tian, et al., Biomaterials 35 (2014) 1618-1626. DOI:10.1016/j.biomaterials.2013.10.077 |
| [53] |
X. Hu, X. Zhao, B. He, et al., Research 2018 (2018) 12. |
| [54] |
Y. Zhu, C. Xu, N. Zhang, et al., Adv. Funct. Mater. 28 (2018) 1706709. DOI:10.1002/adfm.201706709 |
| [55] |
C. Wu, Q. He, A. Zhu, et al., ACS Appl. Mater. Interfaces 6 (2014) 21615-21623. DOI:10.1021/am5066128 |
| [56] |
B. Cao, F. Xiao, D. Xing, X. Hu, Small 14 (2018) 1802008. DOI:10.1002/smll.201802008 |
| [57] |
Y. Yu, F. Bu, H. Zhou, et al., Mater. Chem. Front. (2020), doi: http://dx.doi.org/10.1039/D1030QM00255K.
|
| [58] |
H. Du, Y. Wang, X. Yao, et al., Polym. Chem. 7 (2016) 5620-5624. DOI:10.1039/C6PY01346E |
| [59] |
C. Zhang, Z. Ying, Q. Luo, et al., J. Polym. Sci.:Polym. Chem. Ed. 55 (2017) 2027-2035. DOI:10.1002/pola.28581 |
| [60] |
W. Chin, C. Yang, V.W.L. Ng, et al., Macromolecules 46 (2013) 8797-8807. DOI:10.1021/ma4019685 |
| [61] |
K. Saha, D.F. Moyano, V.M. Rotello, Mater. Horiz. 1 (2014) 102-105. DOI:10.1039/C3MH00075C |
 2020, Vol. 31
2020, Vol. 31