b The Affiliated Hospital of Qingdao University, Qingdao 266003, China
Sulfur-containing species (including organic sulphur, inorganic sulphur and biothiols) play vital roles in varieties of pathological conductions and physiological functions because of their widespread occurrence in living organism [1]. Abnormal expression of sulfur-containing species has been authenticated to be closely associated with certain diseases, such as liver injury, cancer, Alzheimer's disease and growth failure [2-4]. Therefore, sensitive discriminating and quantitative detection of sulfur-containing species not only provide an in-depth insight into the biogeochemical cycle in living systems, but also contribute to the early diagnosis of diseases. Traditional detection techniques such as electrophoresis, atomic absorption spectrometry, mass spectrometry, high performance liquid chromatography and organic fluorescent probes although can realize sensitive sulfur-containing species detection [5-7], limited detection capability and time-consuming techniques still form a major bottleneck for their further application [8]. Thus, developing high-throughput and more convenient methods for sulfur-containing species identification is still highly desirable.
Inspired by human olfactory and gustatory sensing systems, array-based multivariable sensing techniques have been energetically developed as a potentially alternative approach towards high-throughput and simultaneous identification of diverse analytes [9-17]. Unlike traditional "lock-and-key" principle, unique optical "fingerprint" responses for multiple targets can be achieved based on nonspecific rather than challenging specific receptors, making it labor- and time-saving [18]. Such approaches can straightforwardly identify multiple diverse sulfur speciation as well as in biofluids such as urines and serums, which may indicate disease states [19, 20]. Until now, although several sulfur speciation identifcation platforms on the basis of array based strategy have been established, most of them mainly focus on the thiols discrimination. However, sensor arrays capable of simultaneously distinguishing different types of sulfur-containing species (organic sulphur, inorganic sulphur and biothiols) are scarcely reported. In addition, rapid signal responses and simplification of the probe synthesis are also facing great challenges. Therefore, developing a time-saving and convenient method for high-throughput identification of various sulfur-containing species is of great significance for disease early warning.
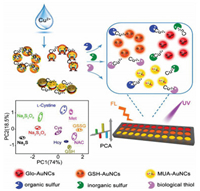
In this work, we developed a triple-channel sensor array for rapid discriminating ten different types of sulfur-containing species based on different fluorescence responses caused from competitive binding between three Au NCs-Cu2+ complexes and different sulfur-containing species. Compared with the traditional sulfur-containing species discrimination sensor arrays, several distinct superiorities of this sensor array make it particularly attractive: (1) Different types of sulfur-containing species including organic sulphur, inorganic sulphur and biothiols can be both simultaneously distinguished. (2) It takes only 2 min to obtain the best response signals. (3) Single or multiple sulfur-containing species in complex biological systems such as urine and serum can be effectively identified. (4) It can not only distinguish serums of cancer patients (such as liver cancer and breast cancer) from that of healthy people, but also distinguish sulfur-oxidizing bacteria (SOB) from sulphur-free bacteria, demonstrating potential prospect for auxiliary clinical diseases warning and point-of-care diagnosis.
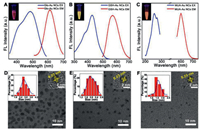
Figs. 1A–C exhibit the excitation (blue line) and emission (red line) spectra of these three kinds of Au NCs. Although these three kinds of Au NCs show different optimal excitation wavelengths, the inset fluorescence photos in Figs. 1A–C demonstrate that they can be both well excited using 365 nm ultraviolet light. TEM results shown in Figs. 1D–F reveal that these Au NCs are well dispersed with an average diameter around 2~3 nm. In addition, remarkable crystalline structure of the Au NCs of the high-resolved lattice planes spacing with 0.23 nm can be clearly observed in the HRTEM image (insets in Figs. 1D–F), which is in consistent with the previous report [21, 22]. As the properties of functional groups on the surface of Au NCs play an important role in analytes identification [23], FT-IR and XPS were carried out for in-depth investigating the surface properties of these three kinds of Au NCs. The asymmetric and symmetric stretching vibration peaks of COO- at 1600 and 1400 cm-1 reveal that there are abundant carboxyl groups on the surface of these three kinds of Au NCs (Figs. S1A-C in Supporting information) [24]. In addition, the binding energies of Au 4f7/2 (around 84.0 eV) and Au 4f5/2 (around 88.0 eV) reveal that both Au(0) and Au(I) exist on the surface of these Au NCs (Figs. S1DF in Supporting information) [25]. It is worth noting that Au(I) surrounded on the surface is reported to be conductive to the stabilization of these Au NCs [26].
|
Download:
|
| Fig. 1. Excitation and emission spectra of (A) Glo-Au NCs, (B) GSH-Au NCs and (C) MUA-Au NCs. The inset shows the corresponding fluorescence photoes under UV light illumination. TEM images of (D) Glo-Au NCs, (E) GSH-Au NCs and (F) MUA-Au NCs. The left and right insets show size distribution and the HRTEM image, respectively. | |
As shown in Scheme 1, this sensor array for sulfur species discrimination consists of three kinds of Au NCs in a 96-well plate. After adding Cu2+ to Au NCs, Cu2+ will act as a bridge to induce the aggregation of Au NCs because of excellent coordination between Cu2+ and abundant carboxyl groups on the surface of these three kinds of Au NCs [27], leading to the aggregation-induced fluorescence quenching. As shown in Figs. 2A–C, all these three kinds of Au NCs aggregated in different degrees after the addition of Cu2+. Owing to the competitive binding, these aggregated Au NCs redispersed after the addition of sulfur-containing species (using GSH as an example, Figs. 2D–F) and resulted in the fluorescence recovery. The TEM results of aggregation and re-dispersion process are consistent with those of DLS results (Fig. S3 in Supporting information). Besides, absorption spectrum was also performed to investigated this process. As shown in Fig. S4 (Supporting information), after the addition of sulfur-containing species (using GSH as an example) to these three AuNCs-Cu2+ complexes, blue shift of absorption peaks can be observed, which further reflects the process of re-dispersion. In details, adding the same sulfur-containing species to these three quenched Au NCs would produce distinct fluorescence recoveries. Meanwhile, adding different sulfur-containing species to the same quenched Au NCs would also produce distinct fluorescence recoveries. Thus, these different cross fluorescence responses can be used as fingerprints for pattern recognition of different sulfur-containing species.
|
Download:
|
| Scheme 1. Design and preparation of the Au NCs-Cu2+ based sensor array for sulfur species discrimination. | |

|
Download:
|
| Fig. 2. TEM images of the (A) Glo-Au NCs, (B) GSH-Au NCs and (C) MUA-Au NCs with the addition of Cu2+, respectively. TEM images of (D) Glo-Au NCs/Cu2+, (E) GSH-Au NCs/Cu2+ and (F) MUA-Au NCs/Cu2+ complex with the addition of GSH, respectively. | |
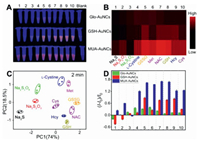
As a proof-of-concept, ten different types of sulfur-containing species including organic sulphur (L-cystine, GSSG, Met), inorganic sulphur (S2-, S2O32-, S2O82-) and biothiols (GSH, NAC, Hcy, and Cys) were selected as the sensing targets. Fig. 3A shows the fluorescence photos of the Au NCs-Cu2+ complexes in the presence of different sulfur-containing species. It can be concluded that each sulfur-containing species shows distinct fluorescence "fingerprint" responses (Figs. 3B and C). Then, classical PCA was performed to transform these pattern response data into linearly uncorrelated principal components. It can be clearly observed from Fig. 3D that these ten different sulfur-containing species were well separated from each other without any overlap, indicating excellent ability for sulfur-containing species discrimination. Most importantly, ten different sulfur-containing species at the concentration of 0.2 μmol/L can still be clearly separated, suggesting its high resolution for sulfur-containing species discrimination (Fig. S2 in Supporting information).
|
Download:
|
| Fig. 3. (A) Fluorescence photos of the Au NCs-Cu2+ complexes in the presence of different sulfur-containing species. (B) Heat map derived from the corresponding fingerprint-like patterns. (C) Fluorescence response of the Au NCs-Cu2+ based sensor array against various sulfur-containing species (500 μmol/L), in which I and I0 represent the fluorescence intensities of Au NCs-Cu2+ complex in the presence and absence of sulfur-containing species, respectively. (D) PCA plot for the discrimination of the sulfur-containing species: 1. Na2S, 2. Na2S2O3, 3. Na2S2O8, 4. L-Cystine, 5. GSSG, 6. Met, 7. NAC, 8. GSH, 9. Hcy, 10. Cys. | |
Here, it is worth noting that the change of fluorescence intensity (I-I0) was directly obtained by photography with the gel bioimaging system instead of fluorescence spectra. Compared with the way of obtaining data by fluorescence spectrum, the biggest advantage of this method is to obtain all the fluorescence change data at the same time without measuring one by one, which is very conducive to the rapid detection. In addition, the reaction conditions including the concentration of Cu2+ and incubation time were optimized. For the optimization of Cu2+, if the concentration of Cu2+ is too low or too high, the space of fluorescence recovery after adding sulfur species is small, which is not beneficial to the array analysis (Fig. S5 in Supporting information). Therefore, 3.0 μmol/L, 1.2 μmol/L and 2.0 μmol/L of Cu2+ were selected for Glo-Au NCs, GSH-Au NCs and MUA-Au NC to form Glo-Au NCs-Cu2+, GSH-Au NCs-Cu2+ and MUA-Au NC-Cu2+ complex, respectively. In addition, as shown in Fig. S6 (Supporting information), these three AuNCs-Cu2+ complexes have good stability, which guarantees the reliability of the subsequent discriminating results. Furthermore, 2 min was selected as the best response time for sulfur-containing species discrimination (Fig. S7 in Supporting information). To the best of our knowledge, this is the fastest method for simultaneously discriminating ten different types of sulfur-containing species reported so far. In addition, the sulfur-containing species had no effect on the fluorescence of these three AuNCs (Fig. S8 in Supporting information).
As is known, it is still great challenges for a sensor array to discriminate different biothiol with similar properties at different concentration levels [28]. Here, the mixtures of GSH, Cys and Hcy with different molar ratios as well as pure GSH, Cys and Hcy were discriminated. As shown in Fig. S9A (Supporting information), these three biothiol with different molar ratios could be well discriminated without overlap. In addition, binary or ternary randomly selected mixtures of sulfur-containing species were also discriminated using this Au NC-Cu2+ based sensor array. As shown in Fig. S9B (Supporting information), these binary or ternary randomly selected mixtures of sulfur-containing species can be well discriminated without any overlap, further demonstrating its potential ability to analyze complicated samples. Moreover, the probability for sulfur-containing species quantification of the Au NC-Cu2+ based sensor array was investigated. Here, PC 1 is simplified to identify the sulfur-containing species since PC 1 is greater than 80%. As shown in Fig. S9C (Supporting information), this Au NC-Cu2+ based sensor array is adequately sensitive to identify sulfur-containing species at micromole concentrations. The linearity of the dose-response curve in Fig. S9D (Supporting information) shows that the interactions are stable and homogeneous, demonstrating it is highly reproducible [29]. Moreover, the accuracy of the established method was evaluated by discriminating unkown samples. As shown in Table S1 (Supporting information), 30 unknown samples could be both accurately identified by blind test, suggesting its good accuracy and remarkable potential for sulfur-containing species discrimination.
To investigate the resolving ability of this Au NC-Cu2+ based sensor array, sulfur-containing species discrimination in the presence of human urine and serum was performed. As shown in Figs. 4A and C, multivariate analysis on the patterns obtained by this Au NC-Cu2+ based sensor array allowed effective identifying single sulfur-containing species in both urine and serum. More importantly, binary or ternary randomly selected mixtures of sulfur-containing species in human urine and serum could also be well discriminated (Figs. 4B and D), further indicating its available potential for sulfur-containing species discrimination in human urine and serum of this Au NC-Cu2+ based sensor array.

|
Download:
|
| Fig. 4. (A, C) PCA plot for discrimination of single sulfur-containing species in human urine and serum, respectively. (a) Na2S, (b) Na2S2O3, (c) Na2S2O8, (d) L-Cystine, (e) GSSG, (f) Met, (g) NAC, (h) GSH, (i) Hcy, (j) Cys. (B, D) PCA plot for discrimination of binary or ternary randomly selected mixtures of sulfur-containing species in human urine and serum, respectively. (a) Na2S + Na2S2O8, (b) Na2S2O8+Na2S2O3, (c) L-Cystine + GSSG, (d) Met + Cys, (e) NAC + GSH, (f) Na2S + Met + GSSG, (g) GSH+Hcy + Cys, (h) Na2S2O8+GSSG + Met, (i) Na2S2O3+-Na2S + L-Cystine and (j) Na2S + L-Cystine +Hcy. | |
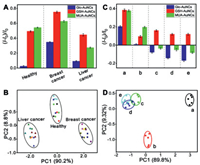
Blood serum-based point-of-care diagnostics provides accurate and rapid diagnosis of disease states [30, 31], consequently, the ability to discriminate serums from cancer patients and healthy people was tested using this Au NC-Cu2+ based sensor array. Here, serum samples from liver cancer and breast cancer have already been definited by the standard method of chemiluminescent microparticle immunoassays in hospital. Compared with serums from healthy people, the content of sulfur-containing species such as inorganic sulphur and biothiols from cancer patients will change to a certain degree. Therefore, serums from liver cancer, breast cancer patients and healthy people will produce different fluorescence responses (Fig. 5A), resulting in the fingerprint PCA identification plot for liver cancer, breast cancer patients and healthy people (Fig. 5B). Moreover, rapid identification of bacterial species contributes to precise drug treatment. Therefore, the ability of this array to discriminate bacteria was also examined. Because the content of sulfur-containing species is different between sulfur-oxidizing bacteria (SOB) and sulfur-free bacteria, different fluorescence responses towards this Au NC-Cu2+ based sensor array can be generated (Fig. 5C). As shown in Fig. 5D, PCA analysis revealed that the SOB and sulfur-free bacteria can also be well discriminated. On the whole, this Au NC-Cu2+ based sensor array not only provides an auxiliary method for breast cancer, liver cancer discrimination, but also for SOB and sulfur-free bacteria, demonstrating its possible prospects in auxiliary clinical diagnose.
|
Download:
|
| Fig. 5. (A) Fluorescence response histogram of these three kinds of Au NCs-Cu2+ against serums from healthy people, breast cancer and liver cancer patients. (B) PCA plot for discrimination of serums from healthy people, breast cancer and liver cancer patients. (C) Fluorescence response histogram of these three kinds of Au NCs-Cu2+ against five kinds of bacteria. (D) PCA plot for discrimination of five kinds of bacteria. (a) Thiobacimonas profunda, (b) Bosea thiooxidans, (c) Aspergillus niger, (d) Staphylococcus albus rosenbach, (e) Escherichia coli (Migula) castellani et chalmers. Bacteria concentrations of OD600 = 0.01. | |
In summary, we developed a convenient sensor array using three kinds of Au NCs-Cu2+ as sensing receptors for rapid identification of different types of sulfur-containing species. Ten different types of sulfur-containing species separately as well as their binary or ternary randomly selected mixtures can be both well discriminated within 2 min. In addition, they can also be achieved in the presence of human urine and serum samples. Furthermore, the most important highlight of this Au NCs-Cu2+ based sensor array is that both rapid serum discrimination (liver cancer, breast cancer and healthy people) and bacteria discrimination (SOB and sulfur-free bacteria) can be realized. We expect, in general, the excellent accuracy and rapid response time will provide a promising approach for point-of-care disease diagnostic.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentsThe authors gratefully acknowledge the support from the National Nature Science Foundation of China (Nos. 21675093, 21505081 and 21974075), the Natural Science Foundation of Shandong Province of China (No. ZR2019YQ13), the Science and Technology Support Plan for Youth Innovation of Colleges and Universities in Shandong Province (No. 2019KJC007) and the Taishan Scholar Program of Shandong Province, China (No. ts20110829).
Appendix A. Supplementary dataSupplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.cclet.2020.04.027.
| [1] |
C.G. Friedrich, F. Bardischewsky, D. Rother, et al., Curr. Opin. Microbiol. 8 (2005) 253-259. DOI:10.1016/j.mib.2005.04.005 |
| [2] |
J. Yin, Y. Kwon, D. Kim, et al., J. Am. Chem. Soc. 136 (2014) 5351-5358. DOI:10.1021/ja412628z |
| [3] |
S.A. Jewell, G. Bellomo, H. Thor, et al., Science 217 (1982) 1257-1259. DOI:10.1126/science.7112127 |
| [4] |
C.X. Yin, K.M. Xiong, F.J. Huo, etal., Angew.Chem.Int.Ed. 56 (2017) 13188-13198. DOI:10.1002/anie.201704084 |
| [5] |
W. Wang, O. Rusin, X. Xu, et al., J. Am. Chem. Soc. 127 (2005) 15949-15958. DOI:10.1021/ja054962n |
| [6] |
P. Liu, Y.Q. Huang, W.J. Cai, et al., Anal. Chem. 86 (2014) 9765-9773. DOI:10.1021/ac5023315 |
| [7] |
S.J. Chen, Y.N. Hong, J.Z. Liu, et al., J. Mater. Chem. B 2 (2014) 3919-3923. |
| [8] |
S. Chen, C.H. Xu, Y.L. Yu, et al., Sens. Actuators B 266 (2018) 553-560. DOI:10.1016/j.snb.2018.03.174 |
| [9] |
S.H. Xu, X. Lu, C.X. Yao, et al., Anal. Chem. 86 (2014) 11634-11639. DOI:10.1021/ac502643s |
| [10] |
S.H. Xu, Z.Z. Su, Z. Zhang, et al., J. Mater. Chem. B 5 (2017) 8748-8753. DOI:10.1039/C7TB02129A |
| [11] |
J.Y. Yang, T. Yang, X.Y. Wang, et al., Anal. Chem. 91 (2019) 6012-6018. DOI:10.1021/acs.analchem.9b00476 |
| [12] |
Y.N. Mao, S.N. Cui, W.T. Li, et al., Sens. Actuators B 296 (2019) 126694. DOI:10.1016/j.snb.2019.126694 |
| [13] |
H. Kong, Y.X. Lu, H. Wang, et al., Anal. Chem. 84 (2012) 4258-4261. DOI:10.1021/ac300718p |
| [14] |
D. Yuan, H.H. Yan, J.H. Liu, et al., Chin. Chem. Lett. 31 (2020) 455-458. DOI:10.1016/j.cclet.2019.07.067 |
| [15] |
Z. Li, S.W. Zhang, T. Yu, et al., Anal. Chem. 91 (2019) 10448-10457. DOI:10.1021/acs.analchem.9b00750 |
| [16] |
L.B. Zheng, P. Qi, D. Zhang, et al., Sens. Actuators B 286 (2019) 206-213. DOI:10.1016/j.snb.2019.01.147 |
| [17] |
T.H. Liu, L.J. Yang, W. Feng, et al., ACS Appl. Mater. Interfaces 12 (2020) 11084-11093. DOI:10.1021/acsami.0c00568 |
| [18] |
F. Pu, X. Ran, J.S. Ren, et al., Chem. Commun. 52 (2016) 3410-3413. DOI:10.1039/C5CC10195F |
| [19] |
S.S. Li, Q.Y. Guan, M.M. Zheng, et al., Chem. Sci. 8 (2017) 7582-7587. DOI:10.1039/C7SC03218H |
| [20] |
S.H. Xu, Y.F. Wu, X.M. Sun, et al., J. Mater. Chem. B 5 (2017) 4207-4213. |
| [21] |
L. Shang, N. Azadfar, F. Stockmar, et al., Small 7 (2011) 2614-2620. DOI:10.1002/smll.201100746 |
| [22] |
X. Zhao, W.T. Li, T. Wu, et al., Anal. Chim. Acta 1079 (2019) 192-199. DOI:10.1016/j.aca.2019.06.056 |
| [23] |
A. Bajaj, O.R. Miranda, I.B. Kim, et al., Proc. Natl. Acad. Sci. U. S. A. 106 (2009) 10912-10916. |
| [24] |
Y. Wang, J.T. Chen, X.P. Yan, Anal. Chem. 85 (2013) 2529-2535. DOI:10.1021/ac303747t |
| [25] |
S.H. Xu, T. Gao, X.Y. Feng, et al., J. Mater. Chem. B 4 (2016) 1270-1275. DOI:10.1039/C5TB02195B |
| [26] |
R.L. Whetten, R.C. Price, Science 318 (2007) 407-408. DOI:10.1126/science.1150176 |
| [27] |
J.R. Bhamore, S. Jha, T.J. Park, et al., Sens. Actuators B 277 (2018) 47-54. DOI:10.1016/j.snb.2018.08.149 |
| [28] |
U.H.F. Bunz, V.M. Rotello, Angew. Chem. Int. Ed. 49 (2010) 3268-3279. DOI:10.1002/anie.200906928 |
| [29] |
H. Pei, J. Li, M. Lv, et al., J. Am. Chem. Soc. 134 (2012) 13843-13849. DOI:10.1021/ja305814u |
| [30] |
S.H. Xu, P.P. Liu, X. Lu, et al., Electrophoresis 35 (2014) 546-553. DOI:10.1002/elps.201300308 |
| [31] |
S. Ray, P.J. Reddy, R. Jain, et al., Proteomics 11 (2011) 2139-2161. DOI:10.1002/pmic.201000460 |
 2020, Vol. 31
2020, Vol. 31 

