b Department of Thoracic Surgery, Qilu Hospital of Shandong University, Ji'nan 250012, China
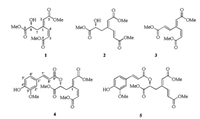
Genus Dracocephalum is an important member of the Lam-iaceae family which comprises more than 60 species of plants. Dracocephalum moldavica L. is an annual herbaceous plant belonging to this genus. In China, wild growing D. moldavica mainly distribute in the northwest area, and also it is cultivated in Xinjiang Autonomous Region. This herb has been believed to possess benefits for the treatment of some kind of chronic diseases including cardiac disease [1-3], antibacterial [4], atherosclerosis [5, 6] and other oxidative-stress related disorders. Previous phytochemical studies revealed that flavonoids [7, 8], triterpenoids [9] and phenylpropanoids [10, 11] are major constituents of D. moldavica. The transcription factor Nrf2 has emerged as the master regulator of a cellular protective mechanism by upregulating antioxidant response element (ARE)-bearing genes encoding antioxidant enzymes, detoxifying enzymes, xenobiotic trans-porters, and stress response proteins. During our ongoing exploration of potent Nrf2 activators, D. moldavica was demon-strated to be a good resource. Subsequent bioassay-guided isolation afforded five new 3, 4-seco-phenylpropanoids, dracomol-phesin A-E (1-5) (Fig. 1). Herein, we report the isolation and structure elucidation of these compounds along with their Nrf2 inducing activity.
|
Download:
|
| Fig. 1. Structures of 1-5 isolated from D. moldavica. | |
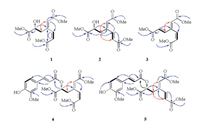
Compound 1 was obtained as yellowish oil. Its molecular formula was determined as C12H16O7 by the analysis of HRESIMS spectrum (m/z 295.0785 [M + Na]+, Calcd. for C12H16O7Na: 295.0794), corresponding to five degrees of unsaturation. The 1H NMR spectrum of 1 showed three olefinic protons at δH 5.88 (m, 1H, H-2), 7.12 (dd, 1H, J = 12.3, 2.0 Hz, H-6) and 5.95 (d, 1H, J =12.3 Hz, H-5); one oxygen-bearing methine at δH 4.32 (dd, 1H, J = 8.3, 4.6 Hz, H-8); a pair of methylene signals at δH 2.98 (m, 1H, H-7a), 2.76 (dd, 1H, J = 14.2, 8.3 Hz, H-7b) and three methoxy groups at δH 3.68 (s, 3H, OMe-3), 3.71 (s, 3H, OMe-4) and 3.74 (s, 3H, OMe-9) (Table S1 in Supporting information). The 13C NMR data together with HSQC spectrum of 1 revealed the existence of 12 carbon signals which could be attributed to three carbonyls (δC 166.0, 166.0 and 173.9), three methoxyls (δC 50.4, 50.6 and 51.1), four olefinics (δC 118.8, 119.4, 144.9 and 152.8), one methine (δC 69.2) and one methylene (δC 42.5). The HMBC correlations from H-6 to C-2 and C-4, H-2 to C-3, H-5 to C-1, as well as OMe-3 to C-3 and OMe-4 to C-4 suggested the presence of a moiety of dimethyl hexa-2, 4-dienedioate (Fig. 2). The 1H-1H COSY correlation of H2-7 with H-8, and the HMBC correlations from H-7 to C-1, C-2, C-6 and C-9, along with OMe-9 to C-9 established the linkage of a methyl 2-hydroxypropionate moiety to C-1. The double bonds between C-1 and C-2, C-5 and C-6 were both Z configuration. NOESY correlation observed from H-7 to H-2 deduced the Z configuration of C-1/C-2 double bond, and a coupling constant (3J) of 12.3 Hz for H-5 and H-6 demonstrated the Z configuration of the double bond between C-5 and C-6.
|
Download:
|
| Fig. 2. Key HMBC (H→C), 1H-1H COSY (-) and NOESY (↔) correlations of 1-5. | |
The absolute configuration of C-8 in compound 1 was determined by using a convenient Mosher esterification method combined with the analysis of circular dichroism (CD) spectrum. For Mosher's method, two portions of 1 were treated with (S)-(+)-and (R)-(-)-α-methoxy-α-(trifluoromethyl) phenylacetyl chloride in deuterated pyridine directly in separate NMR tubes, which afforded the (R)- and (S)-MTPA ester derivatives (1r and 1s, respectively) of 1. The 1H NMR spectra of 1r and 1s were obtained by measuring the reaction tubes directly. The proton signals of diastereomeric MTPA esters (1r and 1s) were clearly different (Figs. S11 and S12 in Supporting information). The observed chemical shift differences (ΔδS-R) was negative on OMe-9, while were positive on H-2, H-5, H-6 and H-7 (Fig. 3), this permitted the assignment of absolute configuration of C-8 to be R [12-14]. Furthermore, the CD spectrum was also measured. As shown in Fig. 3, the negative cotton effect at 222 nm (Δε -4.23, π→π* transition) supported the 8R configuration, which was consistent with reported data in literatures [15, 16]. Thus, the structure of 1 was established and named as dracomolphesin A.

|
Download:
|
| Fig. 3. ΔδS-R values of the MTPA esters of 1 (left), and CD spectra of 1, 2, 4 and 5 (right). | |
Compound 2 was isolated as yellowish oil. Its molecular formula was determined as C12H16O7 on the basis of HRESIMS data (m/z 290.1234 [M + NH4]+; Calcd. for C12H20O7N: 290.1240), which was exactly the same as 1. The 1H NMR and 13C NMR data of 2 showed high degree of resemblance to those of 1, except for the splitting pattern of H-5 and H-6, suggested that the two compounds were isomers with different configuration at C-5/C-6 double bond. The 16.3 Hz coupling constant (3J) for H-5 and H-6 suggested an E configuration of the double bond between C-5 and C-6. NOESY correlations from H-7 to H-2 and H-5 further confirmed the E configuration of C-5/C-6 double bond, also indicated the Z configuration of the double bond between C-1 and C-2. The CD curves of 1 and 2 were closely similar with the same specific Cotton effect at 222 nm, which indicated that the absolute configuration of 2 was identical to 1. Thus, the structure of 2 was assigned and named as dracomolphesin B.
Compound 3 was isolated as yellowish oil. The HRESIMS of 3 exhibited a pseudo molecular ion peak at m/z 255.0863 [M+H]+ (Calcd. for C12H15O6: 255.0869), which was 18 AMU less than that of 1 and 2, and corresponded to a molecular formula of C12H14O6. Compared the NMR data of 3 with those of 1 indicated that 3 has a very similar structure with 1. The difference existed was a methyl acrylate substitution in 3 instead of a methyl 2-hydroxypropionate, this was revealed by signals for H-7 (δH 7.38, dd, J = 15.8, 0.8 Hz) and H-8 (δH 6.09, d, J =15.8 Hz). The configuration of the C-7/C-8 double bond was confirmed as E by the coupling constant of 15.8 Hz for H-7 and H-8. The deduced structure was further confirmed by the analysis of 13C NMR and 2D NMR data.
Compound 4 was obtained as pale-yellow amorphous powder. The molecular formula was determined as C22H24O10 by HRESIMS data (m/z 471.1245 [M + Na]+; Calcd. for C22H24O10Na: 471.1267), suggesting eleven degrees of unsaturation. By analyzing 1D and 2D NMR spectra of 4, the proton and carbon signals were assigned into two structural moieties (Table S2 in Supporting information). The first moiety was the same as compound 1, inferred from the following key signals: three olefinic protons at δH 5.90 (m, 1H, H-2), 7.13 (dd, 1H, J = 12.2, 2.2 Hz, H-6) and 5.98 (d, 1H, J =12.2 Hz, H-5); one oxygen-bearing methine at δH 5.28 (dd, 1H, J = 8.7, 4.5 Hz, H-8); a pair of methylene signals at δH 3.18 (m, 1H, H-7a), 3.05 (dd, 1H, J = 14.5, 8.7 Hz, H-7b) and three methoxy groups at δH 3.67 (s, 3H, OMe-3), 3.71 (s, 3H, OMe-4) and 3.76 (s, 3H, OMe-9). The second moiety was determined as ferulic acyl, deduced from a typical ABX system signals at δH 7.05 (d, 1H, J =1.9 Hz, H-2'), 6.93 (d, 1H, J =8.2 Hz, H-5') and 7.09 (dd, 1H, J = 8.2, 1.9 Hz, H-6'); a pair of olefinic protons at δH 7.67 (d, 1H, J =15.9 Hz, H-7') and 6.33 (d, 1H, J =15.9 Hz, H-8'); a methoxyl at δH 3.94 (3H, s, OMe-3'). The two moieties were linked through the formation of an ester bond between C-8 hydroxyl and ferulic acid, as evidenced by the HMBC correlation from H-8 to C-9'. Detailed analysis of its 2D NMR data confirmed the structure. The configuration of the three double bonds was assigned according to the 3J coupling constant. The absolute configuration of 4 was determined to be identical with that of 1 based on their similar CD spectra, that is, the C-8' carbon was R configuration. Thus, the structure of 4 was assigned and named as dracomolphesin D.
Compound 5 was obtained as pale-yellow amorphous powder with the molecular formula of C22H24O10, as determined by the HRESIMS (m/z 471.1258 [M + Na]+; Calcd. for C22H24O10Na: 471.1267). The 1H NMR and 13C NMR spectra of 5 closely resembled those of 4, the only difference was that the configuration of C-5/C-6 double bond, which was confirmed by the splitting pattern of H-5 and H-6. The absolute configuration of 5 was determined the same as 4 according to their similar CD curves. The structure of 5 was thus clarified and was named as dracomolphesin E.
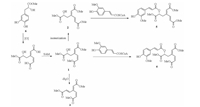
Compounds 1-5 represented an aromatic ring cleaved skeleton of phenylpropanoids. Dioxygenases have been identified from certain members of plant possessing the function of opening aromatic ring [17-20]. We proposed a plausible biogenetic pathway of dracomolphesin A-E (1-5) with salvianic acid A methyl ester (6), a simple phenylpropanoid generated from shikimate pathway, as the biogenetic precursor. Compound 6 was also isolated from D. moldavica with a larger amount than 1-5. As shown in Scheme 1, an intradiol dioxygenase involved oxidative cleavage of the cyclic ring of 6 between C-3 and C-4 produced 3, 4-seco-phenylpropanoids, based on this intermediate, subsequent esterification, isomerization and dimerization may lead to the generation of 1-5.
|
Download:
|
| Scheme 1. Possible biosynthetic pathways of 1-5. | |
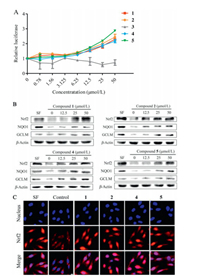
The bioactivity of 1-5 were examined using a stable antioxidant response element (ARE)-dependent reporter gene assay system derived from HEK 293 cells. Compounds 1, 2, 4 and 5 increased the luciferase activity in a dose-dependent manner, suggesting that these compounds were potential Nrf2 transcriptional activators, while compound 3 did not show obvious activity (Fig. 4A). To further determine whether these compounds induced Nrf2 pathway, the protein levels of Nrf2 and its target genes including NQO1 and GCLM were measured by immunoblot analysis. As shown in Fig. 4B, 1, 2, 4 and 5 treatments enhanced both Nrf2 protein expression and the protein levels of NQO1 and GCLM dose-dependently. Immunofluorescence analysis indicated that 1, 2, 4 and 5 trigged Nrf2 translocation from cytoplasm to nucleus (Fig. 4C). 2.5 μmol/L of sulforaphane (SF) was used as positive control.
|
Download:
|
| Fig. 4. Nrf2 inducing activity of compounds 1, 2, 4 and 5. (A) ARE-dependent luciferase assay of compounds 1-5. (B) Compounds 1, 2, 4 and 5 upregulated the protein levels of Nrf2, NQO1 and GCLM. (C) Compounds 1, 2, 4 and 5 activated the nuclear translocation of Nrf2 protein. | |
In conclusion, five 3, 4-seco-phenylpropanoids, dracomolphesin A-E (1-5), were isolated from D. moldavica. These compounds represented an example of phenylpropanoids with an unusual aromatic ring cleaved skeleton. Dioxygenases were proposed to involve in the opening of aromatic ring. Compounds 1, 2, 4 and 5 were identified as novel Nrf2 activators. This study enriched the structural diversity of phenylpropanoids and provided promising target molecules for acquring Nrf2 activators.
AcknowledgmentsThis work was supported by the National Natural Science Foundation of China (No. 81973202) and the Key Research and Development Program of Shandong Province (No. 2018GSF118085).
Appendix A. Supplementary dataSupplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.cclet.2019.09.036.
| [1] |
J. Jiang, X. Yuan, T. Wang, et al., Cardiovasc. Toxicol. 14 (2014) 74-82. DOI:10.1007/s12012-013-9221-3 |
| [2] |
M.E. Tan, C.H. He, W. Jiang, et al., Int. J. Nanomed. 12 (2017) 3253-3265. DOI:10.2147/IJN.S131893 |
| [3] |
C. Zeng, W. Jiang, X. Yang, et al., Sci. Rep. 8 (2018) 17491. DOI:10.1038/s41598-018-35726-4 |
| [4] |
A. Ehsani, O. Alizadeh, M. Hashemi, et al., Vet. Res. Forum 8 (2017) 223-229. |
| [5] |
W. Cao, N. Hu, Phytother. Res. 31 (2017) 1240-1248. DOI:10.1002/ptr.5846 |
| [6] |
J. Xing, K. Peng, W. Cao, et al., Pharm. Biol. 51 (2013) 74-83. DOI:10.3109/13880209.2012.711839 |
| [7] |
A. Sultan, Bahang, H.A. Aisa, Chem. Nat. Compd. 44 (2008) 366-367. DOI:10.1007/s10600-008-9065-4 |
| [8] |
S. Yang, L. Wang, X. Guo, et al., Nat. Prod. Res. 27 (2013) 201-207. DOI:10.1080/14786419.2012.666746 |
| [9] |
Q. Zeng, H.Z. Jin, J.J. Qin, et al., Chem. Biodivers. 7 (2010) 1911-1929. DOI:10.1002/cbdv.200900188 |
| [10] |
Q. Li, Y. Liu, L. Han, et al., J. Sep. Sci. 39 (2016) 4071-4085. DOI:10.1002/jssc.201600645 |
| [11] |
J.L. Zhang, R.J. Yan, N. Yu, et al., Nat. Prod. Res. 32 (2018) 370-373. DOI:10.1080/14786419.2017.1359168 |
| [12] |
Y.Y. Lu, X.P. Gong, Y.B. Xue, et al., Chin. Chem. Lett. 28 (2017) 1460-1464. DOI:10.1016/j.cclet.2017.03.024 |
| [13] |
W.Z. Zeng, S. Que, Q.Y. Zhang, H. Liang, Chin. Chem. Lett. 25 (2014) 1153-1156. DOI:10.1016/j.cclet.2014.05.040 |
| [14] |
X. Zheng, X. Zheng, C. Zhang, et al., Chin. Chem. Lett. 30 (2019) 428-430. DOI:10.1016/j.cclet.2018.09.011 |
| [15] |
Y.N. Shi, Y.M. Shi, L. Yang, et al., J. Ethnopharmacol. 162 (2015) 87-96. DOI:10.1016/j.jep.2014.12.038 |
| [16] |
T. Murata, K. Sasaki, K. Sato, et al., J. Nat. Prod. 72 (2009) 1379-1384. DOI:10.1021/np800781t |
| [17] |
W. Schliemann, U. Steiner, D. Strack, Phytochemistry 49 (1998) 1593-1598. DOI:10.1016/S0031-9422(98)00276-3 |
| [18] |
L. Christinet, F.X. Burdet, M. Zaiko, et al., Plant Physiol. 134 (2004) 265-274. DOI:10.1104/pp.103.031914 |
| [19] |
L.E. Contreras-Llano, M.A. Guerrero-Rubio, J.D. Lozada-Ramírez, et al., mBio 10 (2019) e00345-19. |
| [20] |
R.E. Durán, V. Méndez, L. Rodríguez-Castro, et al., Front. Microbiol. 10 (2019) 528. DOI:10.3389/fmicb.2019.00528 |
 2020, Vol. 31
2020, Vol. 31 

