b University of Chinese Academy of Sciences, Beijing 100049, China;
c Jilin Biomedical Polymers Engineering Laboratory, Changchun 130022, China
Presently, nanomedicine with unique advantages, including excellent water dispersity, selective intratumoral accumulation, controllable release, improved efficacy, reduced side effects, and so forth, attracts increasing attention in the therapy of malignancies [1-6]. However, the unexpected biodistribution and drug release of nanomedicine in the undesired sites always result in severe side effects toward healthy tissue and cells [7]. Therefore, the high-specific drug delivery into the lesion sites is still a significant challenge in cancer therapy.
Several studies reported that the introduction of tumor-specific ligands, e.g., small molecules, peptides, nuclear acid aptamers, and polysaccharides, to the external surface of nanomedicine realizes the efficient targetability [7-9]. The targetability of nanomedicine upregulates drug accumulation at the tumor site and largely avoids non-specific uptake by healthy cells. However, off-target is still a common issue for smart drug delivery.
The introduction of responsiveness to the endogenous and exogenous stimuli into the nanomedicine further improves the selectivity of drug delivery into the tumor cells [10-13]. Among all these stimuli, pH is one of the most promising ones because of the dramatic potentials of pH in different tissues and cellular compartments [14, 15]. For instance, the pH of physiological environments is about 7.4, while in tumor tissue, the value reduces to 6.8. More importantly, the pH in the endosome and lysosome dramatically decreased to 5.0–6.5 [16]. Based on the gradient pH changes, various pH-responsive linkages, such as amide, orthoester, hydrazone, imine, and acetal bonds [16], with different pH-sensitivities, have been applied for the extracellular or intracellular drug delivery [17, 18].
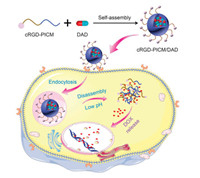
To specifically recognize the tumor cells and selectively deliver the antineoplastic agent into the tumor cells, we developed the targeted pH-responsive polyion complex (PIC) micelle from the positively charged cRGDfC-modified poly(ethylene glycol)-block-poly(L-lysine) (cRGD-PEG-b-PLL) and anionic acidity-activatable doxorubicin (DOX) prodrug (DAD) with the modification of 2, 3-dimethylmaleic anhydride (DMMA), as shown in Scheme 1. The non-pH-responsive PIC micelles with and without cRGDfC from polypeptide and the succinic anhydride (SA)-decorated DOX prodrug (SAD) were prepared as controls to demonstrate the superiority of pH-responsive platform. Compared with PICM/DAD, cRGD-PICM/SAD, and PICM/SAD, cRGD-PICM/DAD acted through a sequential dual-targeting approach. First, cRGD-PICM/DAD selectively accumulated at the tumor site mediated by the ligand-receptor interaction between the cRGDfC on the surface of PIC micelle and the αvβ3 integrin on the membrane of tumor cells. Subsequently, DOX was activated under the intracellular acidic conditions, which resulted in enhanced antitumor efficacy and reduced side effects. Hence, we believe that the targeted PIC micelle is a promising candidate for acidity-mediated intracellular drug release in cancer therapy.
|
Download:
|
| Scheme 1. Schematic illustration of targeted intracellular drug delivery by pH-responsive PIC micelle. | |
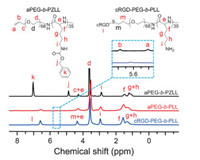
The targeted PIC micelle was prepared by the electrostatic interaction of positively charged cRGD-PEG-b-PLL and negatively charged pH-responsive DAD or non-pH-responsive SAD. cRGD-PEG-b-PLL was synthesized through the sequential ring-opening polymerization, deprotection, and thiol-ene reaction according to the protocol reported in our previous studies (Scheme S1 in Supporting information) [18, 19]. The successful synthesis of cRGD-PEG-b-PLL and its parent polypeptides were demonstrated by the proton nuclear magnetic resonance (1H NMR), as shown in Fig. 1. The characteristic resonances at 4.91 and 3.66 ppm belonged to the protons in the side benzyl group of PZLL (-CH2OC(O)-) and the methylene unit of aPEG (-CH2CH2O-), respectively, demonstrating the successful synthesis of aPEG-b-PZLL. What is more, the degree of polymerization of PLL was calculated to be 35 by 1H NMR spectra. The disappearance of the chemical shifts of proved the successful deprotection and confirmed the synthesis of protons in C6H5CH2- at 7.04 ppm and –CH2OC(O)- at 4.91 ppm proved the successful deprotection and confirmed the synthesis of aPEG-b-PLL. The signal appearance of protons in the phenyl group of cRGD at 7.11 ppm and the disappearance of proton signals in CH2=CH-at 5.70 and 5.49 ppm, indicated the successful synthesis of cRGD-PEG-b-PLL (Fig. 1).
|
Download:
|
| Fig. 1. 1H NMR spectra of aPEG-b-PZLL, aPEG-b-PLL, and cRGD-PEG-b-PLL. | |
The acidity sensitive and insensitive DOX prodrugs DAD and SAD were synthesized by the ring-opening reaction between the amino group in DOX and the corresponding anhydrides DMMA and SA (Scheme S2 in Supporting information). We also confirmed the successful synthesis of DAD and SAD by 1H NMR and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). As shown in Fig. S1 (Supporting information), the peaks appeared at 2.09 and 1.95 ppm were assigned to methyl protons in –C(CH3)=C(CH3) of DAD. Similarly, the characteristic resonance signals of protons in -C(O)CH2CH2C(O)-at 2.50 and 2.34 ppm indicated the successful synthesis of SAD. The data of MALDI-TOF MS further confirmed the successful synthesis of DAD and SAD. The molecular weights of SAD matched the theoretical values, but the molecular weight of DAD was higher than the theoretical value, which was because the DAD bond to Na+ and some DAD-Na+ lost a molecule of H2O. All these results indicated that both the polymers and prodrugs were successfully synthesized.
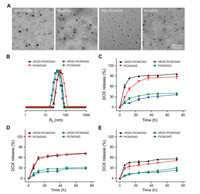
All the PIC micelles, cRGD-PICM/DAD, PICM/DAD, cRGD-PICM/ SAD, and PICM/SAD, were prepared by the electrostatic interaction-incorporated complex of cationic polypeptides and anionic prodrugs[19].The encapsulation efficiencies of all the micelles were about 95.0 wt%, and the drug loading content was about 10.0 wt%. Transmission electron microscopy (TEM) and dynamic laser scattering (DLS) assays were performed to detect the morphologies and sizes of these PIC micelles (Fig. 2). As shown in Fig. 2A, all these PIC micelles self-assembled into spherical nanoparticles with various unified sizes. Typically, the diameters of cRGD-PICM/DAD and PICM/DAD were detected to be 75.4 3.5 and 81.9 5.5 nm by TEM, respectively, which was slightly larger than those of cRGD-PICM/SAD (58.2 4.0 nm) and PICM/SAD (61.6 4.5 nm). What is more, the presence of cRGD on the surface of PIC micelles never affected their morphologies and sizes. In contrast, the sizes from DLS were 97.2 3.5, 109.6 3.8, 80.6 2.3, and 85.8 2.9 nm for the above PIC micelles (Fig. 2B), which were slightly larger than those measured by TEM because the dehydration induced the shrinkage of micelle in the process of sample preparation for TEM detection. The PIC micelles with diameters at around 100 nm are appropriate for the potential selective intratumoral accumulation through the enhanced permeability and retention effect.
|
Download:
|
| Fig. 2. Characterizations of PIC micelles. (A) TEM images and (B) Rhs of cRGD-PICM/DAD, PICM/DAD, cRGD-PICM/SAD, and PICM/SAD. (C–E) In vitro DOX release from cRGD-PICM/DAD, PICM/DAD, cRGD-PICM/SAD, or PICM/SAD at pH 5.5 (C), 6.8 (D), or 7.4 (E). Scale bar: 500 nm. | |
As shown in Figs. 2C–E, the in vitro DOX release behaviors of these PIC micelles were detected in phosphate-buffered saline (PBS) at pH 5.5, 6.8, and 7.4. For the DAD-loaded PIC micelles, the cumulative release of DOX could be accelerated by the decrease of pH from 7.4 to 6.8 and 5.5. Typically, the release percentages of loaded DOX in cRGD-PICM/DAD at pH 7.4, 6.8, and 5.5 were 51.21%, 71.30%, and 95.69%, respectively. The faster release rate of DOX in acidic conditions should be attributed to the more rapid decomposition of DAD than DOX. For the unresponsive systems with payload SAD, the decrease of pH almost unaffected the behaviors of drug release. All these data demonstrated that the DAD-loaded PIC micelle cRGD-PICM/DAD realized effective intracellular drug release.
The uptake behaviors of PIC micelles by HepG2 cells was investigated by confocal laser scanning microscope (CLSM) and flow cytometry (FCM). As shown in Fig. 3A, strong signal of intracellular DOX fluorescence in HepG2 cells after incubation with cRGD-PICM/DAD, PICM/DAD, and free DOX was revealed. In contrast, only slight DOX fluorescence signal was observed in HepG2 cells incubated with cRGD-PICM/SAD or PICM/SAD. What is more, the internalization of DOX by HepG2 cells after incubation with cRGD-PICM/DAD was higher than that of PICM/DAD, which was only slightly lower than that of free DOX, suggesting the targeting PIC micelles could be recognized and internalized by HepG2 cells more effectively. As depicted in Fig. 3B, the quantitative analysis by FCM further proved much higher DOX concentration in the pH-responsive PIC micelles than those in the groups of corresponding non-pH-responsive PIC micelles, especially after coincubation with cRGD-PICM/DAD, owing to the enhanced cell uptake mediated by the ligand-receptor internalization and the faster DOX release from pH-responsive DAD formulations. All results confirmed the enhanced uptake and selective intracellular DOX release of cRGD-PICM/DAD.

|
Download:
|
| Fig. 3. Cell uptake and cytotoxicity of PIC micelles in vitro. (A) Typical CLSM image and(B) FCM profile of HepG2 cells treated with cRGD-PICM/DAD, PICM/DAD, cRGD-PICM/ SAD, PICM/SAD, or DOX for 2 h. (C) In vitro cytotoxicity of cRGD-PICM/DAD, PICM/DAD, cRGD-PICM/SAD, PICM/SAD, and DOX after coincubation for 48 h. Data are represented as mean±SD (n = 3; *P < 0.5, **P < 0.01, ***P < 0.001). Scale bar: 40 μm. | |
A methylthiazol tetrazolium assay detected the in vitro cytotoxicity of PIC micelles toward HepG2 cells. In accordance with the results of cell uptake, the pH-responsive PIC micelles showed greater antitumor efficacy than that of corresponding non-pH-responsive ones at the same DOX concentration, as shown in Fig. 3C. In addition, the introduction of targeting ligand cRGD upregulated the cytotoxicity of PIC micelles. Typically, the cell viability after incubation with cRGD-PICM/DAD or PICM/DAD at the DOX concentration of 2.5 μg/mL was 23.07% and 56.51%, respectively. The increased cytotoxicity of cRGD-PICM/DAD resulted from the effective cell uptake and rapid intracellular DOX release behaviors.
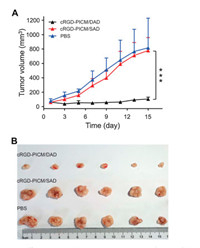
Moreover, the superior antitumor efficacy of cRGD-PICM/DAD in vivo was demonstrated on the xenograft human HepG2 hepato-ma-bearing nude mouse. As expected, cRGD-PICM/DAD at a DOX equivalent dose of 5.0 mg per kg body weight exhibited the best therapeutic effect, showing the smallest tumor volume in comparison with cRGD-PICM/SAD and PBS (Fig. 4). Because of the limited release of DOX and low activity of SAD, cRGD-PICM/SAD exhibited comparable antitumor efficacy as negative control PBS. Such an excellent therapeutic effect of cRGD-PICM/DAD over cRGD-PICM/SAD is consistent with the antitumor results in vitro, owing to the effective recognition by HepG2 cells and the acidity-triggered DOX release. In addition, it should be noted that the mice injected with cRGD-PICM/DAD showed no noticeable decrease in body weight (Fig. S2 in Supporting information) and no toxicity to other organs, such as the heart, liver, spleen, lung, and kidney (Fig. S3 in Supporting information). Hence, the targeted acidity-sensitive PIC micelle has the potential to provide a promising platform for intracellular drug delivery in enhanced cancer treatment.
|
Download:
|
| Fig. 4. Antitumor efficacies in vivo. (A) Tumor volume changes of xenograft human HepG2 hepatoma model during treatment with cRGD-PICM/DAD, cRGD-PICM/SAD, or PBS. (B) Photographs of tumors after different treatments. Data are represented as mean±SD (n = 6; *P < 0.5, **P < 0.01, ***P < 0.001). | |
In summary, the targeted and acidity-sensitive cRGD-PICM/ DAD was prepared by the electrostatic interaction between cRGD-PEG-b-PLL and DAD. The spherical cRGD-PICM/DAD with a diameter of about 100 nm exhibited the increased cell uptake and acidity-responsive intracellular drug release compared with the non-targeted and/or non-acidity-activatable controls.Furthermore, cRGD-PICM/DAD showed the best inhibition efficacy toward hepatoma in vitro and in vivo. Therefore, the smart, targeted PIC micelles with enhanced antitumor efficacy and decreased side effects show great potential in cancer therapy.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentsThis research was financially supported by the National Natural Science Foundation of China (Nos. 51973216, 51873207, 51833010, 51703225, 51673190, 51673187, 51603204 and 51520105004), the Science and Technology Development Program of Jilin Province (No. 20190201068JC), the National Key Research and Development Program of China (No. 2016YFC1100701), the Youth Talents Promotion Project of Jilin Province (No. 181909), and the Youth Innovation Promotion Association of Chinese Academy of Sciences (No. 2019005).
Appendix A. Supplementary dataSupplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.cclet.2019.12.001.
| [1] |
J. Wang, W. Xu, S. Li, et al., J. Biomed. Nanotechnol. 14 (2018) 2102-2113. DOI:10.1166/jbn.2018.2624 |
| [2] |
J. Wolfram, M. Ferrari, Nano Today 25 (2019) 85-98. DOI:10.1016/j.nantod.2019.02.005 |
| [3] |
Q. Wang, P. Zhang, Z. Li, et al., Theranostics 9 (2019) 1426-1452. DOI:10.7150/thno.31683 |
| [4] |
X.R. Feng, J.X. Ding, R. Gref, X.S. Chen, Chin. J. Polym. Sci. 35 (2017) 693-699. DOI:10.1007/s10118-017-1932-7 |
| [5] |
C. Yao, J. Tian, H. Wang, et al., Chin. Chem. Lett. 28 (2017) 893-899. DOI:10.1016/j.cclet.2017.01.005 |
| [6] |
J. Peng, T. Qi, J. Liao, et al., Theranostics 4 (2014) 678-692. DOI:10.7150/thno.7869 |
| [7] |
S. Zhou, D. Wu, X. Yin, a et, J. Exp. Clin. Cancer Res 36 (2017) 24. DOI:10.1186/s13046-017-0492-6 |
| [8] |
L.C. Wyatt, A. Moshnikova, T. Crawford, et al., Proc. Natl. Acad. Sci. U. S. A 115 (2018) E2811-E2818. DOI:10.1073/pnas.1715350115 |
| [9] |
Q. Yang, J. Peng, Y. Xiao, et al., ACS Appl. Mater. Interfaces 10 (2018) 150-164. DOI:10.1021/acsami.7b14705 |
| [10] |
J. Chen, J. Ding, Y. Wang, et al., Adv. Mater 29 (2017) 1701170. DOI:10.1002/adma.201701170 |
| [11] |
W. Xu, J. Ding, X. Chen, Biomacromolecules 18 (2017) 3291-3301. DOI:10.1021/acs.biomac.7b00950 |
| [12] |
Y. Zhang, J. Zhang, W. Xu, et al., Acta Biomater. 77 (2018) 63-73. DOI:10.1016/j.actbio.2018.07.021 |
| [13] |
J. Chen, J. Ding, C. Xiao, X. Zhuang, X. Chen, Biomater. Sci. 3 (2015) 988-1001. DOI:10.1039/C5BM00044K |
| [14] |
J. Wang, W. Xu, H. Guo, et al., Colloids Surfaces B 135 (2015) 283-290. DOI:10.1016/j.colsurfb.2015.07.065 |
| [15] |
Y. Zhang, L. Chen, J. Ding, a et, J. Control. Release 213 (2015) e135-e136. |
| [16] |
Q. Lei, S.B. Wang, J.J. Hu, et al., ACS Nano 11 (2017) 7201-7214. DOI:10.1021/acsnano.7b03088 |
| [17] |
Y. Song, D. Li, J. He, M. Zhang, P. Ni, Chin. Chem. Lett. 30 (2019) 2027-2031. DOI:10.1016/j.cclet.2019.04.052 |
| [18] |
J. Ding, D. Li, X. Zhuang, X. Chen, Macromol. Biosci. 13 (2013) 1300-1307. DOI:10.1002/mabi.201300160 |
| [19] |
J. Chen, J. Ding, Y. Zhang, et al., Polym. Chem. 6 (2015) 397-405. DOI:10.1039/C4PY01149J |
 2020, Vol. 31
2020, Vol. 31 

