Inflammation is the pathophysiological response of body to harmful stimuli, including pathogens, cell injury and chemicals [1-3]. It may lead to serious tissue destruction in some cases, and chronic inflammation may lead to many diseases, such as atherosclerosis, hay fever, and even cancer [4-6]. However, for most situations, inflammation is a protective response involving the cooperation of immune cells, blood vessels and chemical mediators [7-9]. As nonspecific immune cells, macrophages are important defenders to eliminate harmful stimuli and end inflammation as soon as possible. Studies have demonstrated and visualized that the defense mechanism of macrophages showed close relation to endogenous hypochlorous acid, nitric oxide, hydrogen peroxide and other ROS/RNS [10-14]. To deepen our understanding of inflammation, many other micro-environmental factors in macrophages need research as well, such as pH. In order to promote activities of enzymes to degrade stimuli, pH in lysosomes of macrophages must be regulated accordingly. It is therefore important to know about the regulating pattern of pH in macrophages, especially in lysosomes.
Numerous pH probes targeting in lysosome were reported in the past few years [15-22]. Most of these pH probes are based on fluorescence "off-on" mechanism that are not suitable for quantify pH value. A few probes based on fluorescence ratio provide opportunity for quantitative monitor pH in living cells. Zhang et al. reported a pH probe of lysosome by linked to a naphthalimide to rhodamine to quantify pH ranging from 4.5 to 5.5 [23]. Han's et al. designed FRET dyad by conjugating rhodamine to coumarin and obtaining relative quantitative pH by using two excitation lasers [24]. Ratiometric sensing of pH on living cells based on nanostructures also display promising properties [25-27]. As we expected, an ideal probe to indicate pH in lysosome would (i) target lysosome, (ii) show "always-on" fluorescence to track lysosome continuously, (iii) have wide response range of pH (4.0–8.0), (iv) indicate pH ratiometrically through single laser excitation, and (v) show minimal background signal.
FRET (Fӧrster resonance energy transfer) system is an ideal candidate for designing probes with fluorescence ratio response. According to our former research, cassettes based on a BODIPY donor and a rhodamine acceptor had been proved tobe a suitableplatformto design probes [28, 29]. This fluorophore pair displayed photo-physical advantages, including high molar extinction coefficient, narrow spectrum, and suitable spectrum overlap to facilitate fluorescence ratio imaging [30]. Therefore, in this design, a BODIPY derivative (BDP) was selected as the pH insensitive donor, and a reduced rhodamine B (RhB) was chosen as the pH sensitive acceptor. Different from our former designs, that conjugated fluorophores through rigid spacer to gain a maximal FRET efficiency of almost 100%, in this design, we linked these two fluorohpores through flexible chain to "tune down" the FRET efficiency. The remaining fluorescence from BDP could be used as an "always-on" tracker of lysosomes (Fig. 1). Moreover, as shown in Fig. 1, the tertiary-amine in RhB enable lysosome selectivity of BDP-RhB due to alkaline effect.

|
Download:
|
| Fig. 1. The structures of BODIPY-rhodamine cassettes through rigid spacer in former study, and BDP-RhB through flexible liker in this study. | |
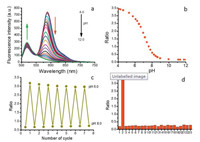
In order to assess the sensitivity of BDP-RhB towards pH, we first studied spectral properties of the probe in solution. As shown in Figs. S1 and S2 (Supporting information), the maximum absorption wavelength of BDP and RhB moiety is 503 nm and 568 nm respectively, and the maximum fluorescence wavelength is 518 nm and 582 nm respectively. pH titration from 12.0 to 4.0 showed that the fluorescence intensity of RhB moiety increased significantly, whereas the fluorescence intensity of BDP moiety decreased slightly (Fig. 2a). Accordingly, as shown in Fig. 2b, the ratio between two fluorescence intensities (I582/I518) showed a significant increase from 0.6 to 3.4 at pH range from 8.0 to 4.0 covering the pH windows of lysosome (pH 4.5–6.5) and cytoplasm (6.5–7.5). The p Ka of the probe was calculated to be 7.1. Furtherly, pH circulating titration at pH 6.0 and 8.0 showed that fluorescence ratio (I582/I518) was stable (Fig. 2c), which revealed that the probe was able to detected pH reversibly and sustainably. In addition, pH response of BDP-RhB showed negligible dependence on diverse metal ions according to selectivity assay including ions, common anions and amino acids (Fig. 2d). And according to reported method [30], the FRET efficiency was calculated to be 95% at pH 4.0 (Supporting information), which is lower than our former report (up to 100%). Therefore, spectral studies indicate our probe has favorable properties for detecting lysosomal pH.
|
Download:
|
| Fig. 2. (a) Fluorescence spectra of BDP-RhB (1 μmol/L) at different pH values. (b) The ratio between fluorescence intensity I582/I518 vs. pH values. (c) Circulating of fluorescence ratio at pH 6.0 and 8.0. (d) The responses of fluorescence ratio (I582/I518) against diverse ions: 1, blank; 2, H+; 3, NH4+; 4, Fe2+; 5, Zn2+; 6, Cu2+; 7, Ca2+; 8, Fe3+; 9, Ag+; 10, K+; 11, Cys; 12, Leu; 13, Arg; 14, Asp; 15, NO3-; 16, Br-; 17, ClO3-; 18, AcO-; 19, SO32-; 20, ClO4-; 21, I-; 22, N3-; 23, H2O2. Excited with 480 nm laser. | |
To further research the pH responding properties of BDP-RhB, we proceeded mechanism studies, including 1H NMR test and density functional theory (DFT) calculation. We compared 1H NMR in CDCl3 before and after adding TFA. As shown in Fig. S3 (Supporting information), before adding TFA, partial open-ring probes result in weak signals at 7.71 and 7.52. While, after adding TFA, signals at 7.71 and 7.52 get stronger, and furthermore, new signals appear at 7.93 and 7.00 and the signal at 6.40 disappears. To investigate the further insights into the electronic structure of BDP-RhB and its protonated form BDP-RhB+2H, the geometry was optimized by DFT calculations at B3LYP/6-31G(d) level. As shown in Fig. S4 (Supporting information), when rhodamine is in "OFF" state, the electron density distribution of the highest occupied molecular orbital (HOMO) is mainly located on the half unit of rhodamine, while the lowest unoccupied molecular orbital (LUMO) is essentially distributed on the whole BODIPY unit.However, for the protonated compound BDPRhB+2H, which is in "ON" state, the HOMO is mainly located on the BODIPY unit, whereas the LUMO is completely located on the whole rhodamine unit.Moreover, the HOMO-LUMO energy gap of BDP-RhB+2H (1.09 eV) is smaller than that of BDP-RhB (1.64 eV), indicating that the absorption onset has occurred a remarkable bathochromic shift to the long wavelength region, which is in accordance with the absorption spectra of the "ON" state rhodamine unit. These theoretical results are well consistent with the experimental data. Therefore, 1H NMR and DFT calculations support the open-ring mechanism in Fig. 1
Next, we evaluated the ability of BDP-RhB to stain live cells and target lysosome. We performed MTT assay before cells imaging, and found the probe showed low cytotoxicity, as cells showed 86.7% viability upon 24 h incubation with 5 μmol/L of BDP-RhB that is 2.5-fold of working concentration (2 μmol/L) (Fig. S5 in Supporting information). BDP-RhB was then proved to stain macrophages seeded in culture dish within 5 min. As shown in Figs. 3a and b, both BDP and RhB channels showed intense fluorescence. Fluorescence in BDP channel is from the "always-on" BDP moiety, while fluorescence in RhB channel is from open-ring RhB due to the acidic microenvironment. Therefore, the ratio between RhB channel and BDP channels can be used for indicating the local pH. Colocalization study of the probe with Lyso-NIR [31], a NIR lysosome tracker, displayed good colocalization with an overlay factor of 0.83 (Figs. 3c–e). As shown in Fig. 3f, intensity profile of BDP channel and Lyso-NIR channel through the region of interest showed high synchronization, demonstrating that our probe can specially localize in lysosomes of macrophage. Further co-stain study with Mito-tracker Deep Red proved that BDP-RhB did not localize in mitochondria (Fig. S6 in Supporting information). The LogP value of the probe is determined to be 1.28 as shown in Fig. S7 (Supporting information), which is in favor of membrane permeability according to predicting theory by Horobin et al. [32].

|
Download:
|
| Fig. 3. Confocal images of macrophages co-stained with BDP-RhB (2 μmol/L) and Lyso-NIR (2 μmol/L). (a) BDP channel; (b) Lyso-NIR channel; (c) bright field; (d) merged image of BDP channel and Lyso-NIR channel; (e) enlarged image; (f) intensity profiles of BDP and Lyso-NIR channels through ROI 1. | |
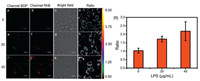
Then we use BDP-RhB to study the variation pattern of pH in an inflammation model of macrophages. We first stimulated macrophages with diverse concentrations of lipopolysaccharide (LPS, 0, 20, 40 μg/mL) for 12 h to induce inflammation, and then stained these cells with BDP-RhB for fluorescence imaging. As shown in Fig. 4, the red fluorescence of RhB moiety increased significantly as the concentration of LPS increased from 0 to 40 μg/mL. Accordingly, the ratio (I582/I518) increased from 1.03 to 2.18 (Fig. 4m and Table S1 in Supporting information), which indicated a significant pH decrease during inflammation.
|
Download:
|
| Fig. 4. Fluorescnce and fluorescence ratio imaging of inflammation macrophage cells stained with BDP-RhB. (a, e, i) BDP channel, (b, f, j) RhB channel, (d, h, l) ratio image between BDP and RhB channels, (c, g, k) bright field image. (m) Histogram of ratio against the concentrations of LPS. Scale bar: 1 μm. | |
In summary, we had developed a BODIPY-rhodamine based ratiometric probe that had a wide pH response range from pH 4.0 to 8.0. The probe had favorable lysosome targeting ability and ratiometric pH response without interference by other ions. Appling these features, we were able to found a decrease of lysosomal pH of macrophages during inflammation.
Declaration of competing interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgmentsThis work was supported by the National Natural Science Foundation of China (Nos. 21421005, 21576040 and 21776037), the Fundamental Research Funds for the Central Universities (No. DUT18RC(3)027) and Supercomputing Center of Dalian University of Technology.
Appendix A. Supplementary dataSupplementary material related to this article canbefound, in the online version, at doi:https://doi.org/10.1016/j.cclet.2019.10.025.
| [1] |
P. Libby, P.M. Ridker, A. Maseri, Circulation 105 (2002) 1135-1143. DOI:10.1161/hc0902.104353 |
| [2] |
O. Takeuchi, S. Akira, Cell 140 (2010) 805-820. DOI:10.1016/j.cell.2010.01.022 |
| [3] |
V. Anttila, H. Stefansson, M. Kallela, et al., Nat. Genet. 42 (2010) 869-873. DOI:10.1038/ng.652 |
| [4] |
F. Balkwill, A. Mantovani, Lancet 357 (2001) 539-545. DOI:10.1016/S0140-6736(00)04046-0 |
| [5] |
A. Mantovani, P. Allavena, A. Sica, et al., Nature 454 (2008) 436-444. DOI:10.1038/nature07205 |
| [6] |
S.J. Galli, M. Tsai, A.M. Piliponsky, Nature 454 (2008) 445-454. DOI:10.1038/nature07204 |
| [7] |
R. Dantzer, J.C. O'Connor, G.G. Freund, et al., Nat. Rev. Neurosci. 9 (2008) 46-56. DOI:10.1038/nrn2297 |
| [8] |
H. Park, Z. Li, X.O. Yang, et al., Nat. Immunol. 6 (2005) 1133-1141. DOI:10.1038/ni1261 |
| [9] |
C.L. Langrish, Y. Chen, W.M. Blumenschein, et al., J. Exp. Med. 201 (2005) 233-240. DOI:10.1084/jem.20041257 |
| [10] |
M.A. Islam, M. Proll, M. Holker, et al., Innate Immun. 19 (2013) 631-643. DOI:10.1177/1753425913477166 |
| [11] |
P. Zhang, H. Wang, Y. Hong, et al., Biosens. Bioelectron. 99 (2018) 318-324. DOI:10.1016/j.bios.2017.08.001 |
| [12] |
Q.W. Xie, R. Whisnant, C. Nathan, J. Exp. Med. 177 (1993) 1779-1784. DOI:10.1084/jem.177.6.1779 |
| [13] |
S. Pawate, Q. Shen, F. Fan, et al., J. Neurosci. Res. 77 (2004) 540-551. DOI:10.1002/jnr.20180 |
| [14] |
C.F. Nathan, J. Clin. Invest. 80 (1987) 1550-1560. DOI:10.1172/JCI113241 |
| [15] |
Y.J. Gong, Z.Z. Kong, M.L. Zhang, et al., Talanta 203 (2019) 1-8. DOI:10.1016/j.talanta.2019.05.037 |
| [16] |
S. Xia, M. Fang, J. Wang, et al., Sens. Actuators B:Chem. 294 (2019) 1-13. |
| [17] |
J. Li, X. Li, J. Jia, et al., Dye Pigment 166 (2019) 433-442. DOI:10.1016/j.dyepig.2019.03.060 |
| [18] |
H. Xu, S. Ma, Q. Liu, et al., Chem. Commun. (Camb.) 55 (2019) 7053-7056. DOI:10.1039/C9CC02481F |
| [19] |
B. Dong, X. Song, C. Wang, et al., Anal. Chem. 88 (2016) 4085-4091. DOI:10.1021/acs.analchem.6b00422 |
| [20] |
J.R. Hou, D. Jin, B. Chen, et al., Chin. Chem. Lett. 28 (2017) 1681-1687. DOI:10.1016/j.cclet.2017.03.037 |
| [21] |
J. Wen, P. Xia, Z. Zheng, et al., Chin. Chem. Lett. 28 (2017) 2005-2008. DOI:10.1016/j.cclet.2017.09.014 |
| [22] |
A.M. Luo, Y. Shao, K.J. Zhang, et al., Chin. Chem. Lett. 28 (2017) 2009-2013. DOI:10.1016/j.cclet.2017.08.037 |
| [23] |
X.F. Zhang, T. Zhang, S.L. Shen, et al., J. Mater. Chem. B 3 (2015) 3260-3266. DOI:10.1039/C4TB02082K |
| [24] |
Z. Xue, H. Zhao, J. Liu, et al., ACS Sens. 2 (2017) 436-442. DOI:10.1021/acssensors.7b00035 |
| [25] |
H. Li, H. Dong, M. Yu, et al., Anal. Chem. 89 (2017) 8863-8869. DOI:10.1021/acs.analchem.7b01324 |
| [26] |
Y. He, Z. Li, Q. Jia, et al., Chin. Chem. Lett. 28 (2017) 1969-1974. DOI:10.1016/j.cclet.2017.07.027 |
| [27] |
R. Arppe, T. Näreoja, S. Nylund, et al., Nanoscale 6 (2014) 6837-6843. DOI:10.1039/C4NR00461B |
| [28] |
X. Zhang, Y. Xiao, X. Qian, Angew. Chem. Int. Ed. 47 (2008) 8025-8029. DOI:10.1002/anie.200803246 |
| [29] |
B.X. Shen, Y. Qian, Z.Q. Qi, et al., J. Mater. Chem. B 5 (2017) 5854-5861. DOI:10.1039/C7TB01344B |
| [30] |
H. Yu, Y. Xiao, H. Guo, et al., Chem.-Eur. J. 17 (2011) 3179-3191. DOI:10.1002/chem.201002498 |
| [31] |
X. Zhang, C. Wang, Z. Han, et al., ACS Appl. Mater. Interfaces 6 (2014) 21669-21676. DOI:10.1021/am506750m |
| [32] |
R.W. Horobin, F. Rashiddoubell, Biotech. Histochem. 88 (2013) 461-476. DOI:10.3109/10520295.2013.780635 |
 2020, Vol. 31
2020, Vol. 31 

