b Quality Management Department, Wuhan Hiteck Biological Pharma Co., Ltd., Wuhan 430056, China
Thiophonate-methyl (TPM) is one of fungicides and pesticides used widely in agricultural production [1] and easy to be metabolized into harmful benzimidazole (BZD) compounds in vivo of plant [2]. Once TPM is sprayed onto plants, TPM and its BZD metabolites will remain in related agricultural products [3], which poses a serious threat to human health [4]. Therefore, it is necessary to monitor the residues of TPM and its BZD metabolites in related agricultural products [5].
However, some unknown harmful TPM's BZD metabolites may not be detected even if they actually remain in agricultural products due to their extremely low concentrations in agricultural products and the complexity of their matrices. In this case, BZD metabolites need to be further identified and then quantified to improve the food safety.
HPLC coupled with mass spectrometry method (HPLC-MS) is a powerful technique for the identification and quantification of individual unknown BZD metabolites [6-9]. The unavailability of BZD metabolite standards and the absence of MS/MS spectra for library or literature matching pose great challenges for identifying the unknown BZD metabolites from complex matrix samples [10]. In recent years, several HPLC-MS/MS methods have been intro-duced for the analysis of some BZD veterinary drugs metabolites in different biological samples such as laying hen, lactating goats, and mouse plasma [10-13]. By contrast, the identification of the unknown BZD metabolites from pesticide has been less reported.
Since all the BZD compounds possess benzimidazole skeleton and then produce similar fragments by MS/MS, the unknown BZD metabolites may be screened out using high-performance liquid chromatography coupled with precursor ion scan-mass spectrom-etry (HPLC-PIS-MS). As for the low-content BZD metabolites in complex biological matrix, sample pretreatment such as solid-phase extraction (SPE) can be adopted [14].
As catalyst and separation media, nickel-based materials have been applied in various fields [14-17]. Since divalent Ni ion in NiO has 6 vacancies which can bind reversibly with the electron-donating imidazole groups, NiO is supposed to have selectivity for the extraction of the analytes with benzimida-zoles. Our previous study has confirmed the good selectivity of homemade SiO2@NiO for the extraction of benzimidazoles in fruits and vegetables [14].
In this study, SiO2@NiO was used as SPE sorbent for the selective extraction of TPM's BZD metabolites. The obtained extract was injected into HPLC-PIS-MS to screen out the potential BZD metabolites. Lastly, high-resolution mass spectrometry (HR-MS/MS) was used to identify the unknown TPM's BZD metabolites in celery cabbage. Subsequently, a quantitative method of the identified metabolites was developed. In addition, the use of TPM and the metabolic time of TPM for food safety were investigated, based on the residue profiles of TPM's BZD metabolites in celery cabbage sprayed with TPM.
The details of the experiment were described in Supporting information. The structures of TPM and its three known BZD metabolites were shown in Fig. S1 (Supporting information). The parameters of instrument were displayed in detail in Tables S1–S3 (Supporting information).
The extraction recoveries of BZDs in celery cabbage samples spiked with three TPM's known BZD metabolites standards at the concentration of 50 ng/mL were evaluated. As shown in Table S4 (Supporting information), the recoveries of BZDs ranged from 79.5% to 83.7%, which indicated that SiO2@NiO SPE could extract the BZD compounds from celery cabbage samples.
Since all BZD compounds have benzimidazole ring structures, they can produce similar characteristic fragment ions in HPLC-MS. In the MS/MS analysis, in order to screen out the similar structural fragments of BZD compounds, the characteristic fragment ions were obtained from the known TPM's BZD metabolites standards by product ion scan mode. The obtained product fragment ions were then used as characteristic ions in the precursor ion scan mode to speculate the precursor ions of the potential metabolites. At last, the empirical formula of compounds was speculated by HR-MS/MS to elucidate the chemical structure of the potential metabolites. Based on the above high-resolution MS data, fragmentation paths can be simulated by Mass Frontier Server Manager 2.0 software.
In order to discover more BZD metabolites of TPM, the characteristic product ions were selected and collision energy (CE) of the precursor ion scan mode was optimized as well.
As shown in Fig. S2 (Supporting information), the known TPM's BZD metabolites produced four characteristic product ions, namely, m/z 160, 132, 92, and 65 in product ion scan mode. Considering the serious interference around the product ion m/z 65 in mass spectrometry, m/z 160, 132, 92 were selected as characteristic product ions.
The collision energy was optimized in PIS mode, as shown in Fig. S3 (Supporting information). When collision energy increased from 20 V to 40 V, all BZD metabolites could be detected, and the chromatographic intensities of BZD compounds increased gradu-ally from 20 V to 30 V, and then reached a plateau after 30 V. Therefore, the collision energy of PIS mode was set as 30 V.
As mentioned above, characteristic product ions m/z 160, 132, and 92 were used for screening the potential metabolites of TPM by the method of HPLC-PIS-MS. To validate the feasibility of this method, the standard solution of three BZDs mixture was spiked to the blank celery cabbage samples and then the resultant samples were analyzed by HPLC-PIS-MS. As shown in Fig. S4 (Supporting information), the three corresponding BZDs could be detected in HPLC-PIS-MS mode. For HPLC-PIS-MS, the narrower its scanning mass width is, the higher its sensitivity will be. However, it will require more time and more celery cabbage samples when the scanning mass width is set to be narrower. Considering the limited sample amount and the analysis time, the scanning mass width of each event was set as 250 Da. In HPLC-PIS-MS analysis, each sample was injected twice with different scanning mass width to monitor each characteristic fragment ion.
Fig. 1 showed the total ion chromatograms (TICs) of celery cabbage samples sprayed with TPM in HPLC-PIS-MS analysis. By comparing the precursor ions of positive celery cabbage samples with those of TICs of blank samples, 21 potential metabolites were found in the positive celery cabbage samples, as shown in Table S5 (Supporting information).

|
Download:
|
| Fig. 1. Total ion chromatograms (TIC) of positive celery cabbage samples. A1 and A2: Characteristic fragment ions m/z 160; B1 and B2: Characteristic fragment ions m/z 132; C1 and C2: Characteristic fragment ions m/z 92. The m/z scan was set ranging from 150 Da to 450 Da (A1, B1, C1) and from 450 Da to 600 Da (A2, B2, C2). | |
Afterwards, the structures of these 21 detected potential BZD metabolites were further examined by high resolution mass spectrometry analysis (full scan and MS/MS).
Of these 21 potential BZD metabolites, three metabolites were identified by comparing the chromatograms and mass spectra of pure standards with those of positive samples. The mass spectra of the BZD standard compounds and the actual positive samples according to the corresponding retention time of the BZD standard compounds were shown in Fig. 2. The mass spectra (HR-MS/MS) of BZDs of positive samples were in correspondence with those of the BZD standard compounds.

|
Download:
|
| Fig. 2. Fragmentation patterns of the known BZD metabolites of TPM in celery cabbage compared with those of the standards. The upper halves (red line) of each mass spectrum displayed the known BZD metabolites of TPM in celery cabbage and the lower halves (blue line) were the authentic standard in MBC (A), 2-NH2BZ (B) and BZ (C). | |
Due to the lack of the corresponding standards and MS/MS spectra for library matching, the chemical structures of other 18 potential BZD metabolites were speculated by HR-MS/MS spectra. Characteristic fragment ions on HR-MS/MS spectra were searched firstly. Afterwards, MS/MS spectra of unknown metabolites were compared with those of known BZD compounds to reveal their differences so as to predict the most possible structures of the unknown BZD metabolites. At last, Mass Frontier Server Manager 2.0 software (Thermo Fisher Scientific, USA) was used to validate the deductive metabolite structures.
Fig. 3 showed the MS2 spectrum of a potential metabolite named M-1 with molecular ion m/z of 354.1496 at the retention time of 19.15 min. M-1 had a fragment ion m/z 160.0489 corresponding to the m/z 160.0486 (C8H6N3O), indicating that M-1 was a BZD compound. In the MS2 spectrum, the exact mass m/z 354.1496 of M-1 was speculated as the elemental composition of C15H18N3O7 and the dominant fragment ion at m/z 192.0755 (C9H9N3O2) derived from the neutral loss of a glucuronic acid group (C6H7O6) of molecular ion at m/z of 354.1496. Herein, the fragmentation pathway of M-1 was diagrammed in Fig. 3 and this metabolite was considered to be the glucuronic acid conjugates of carbendazim for the first time.

|
Download:
|
| Fig. 3. MS/MS spectrum and the fragmentation pathway of TPM's BZD metabolites. | |
In total, this study identified four BZD metabolites. Of them, one metabolite was identified for the first time. In addition to the qualitative analysis of TPM's BZD metabolites, this study also examined their metabolism and their residue quantity. Based on the extraction condition reported in our previous paper [18] and HR-MS/MS analysis, the analytical method of TPM's BZD metab-olites in celery cabbage were evaluated through linearity, reproducibility, recovery, the limit of detection (LOD) and the limit of quantification (LOQ).
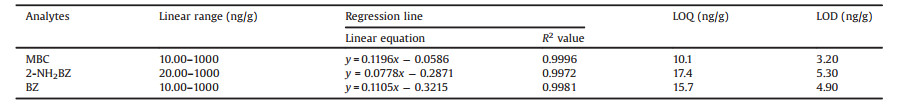
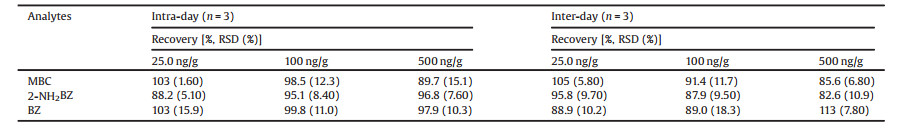
The calibration curves were constructed by the mean peak area ratios of three TPM's BZD metabolite standards to IS versus the spiked concentrations in celery cabbage samples. The least square was used to assess the linearity of calibration curves. Their linearity, LODs, and LOQs were listed in Table 1. A good linearity within the range of 10.00–1000 ng/g was exhibited with R2 from 0.9972 to 0.9996 for the three TPM's BZD metabolite standards. The LODs and LOQs were calculated on the basis of the concentrations at which signal-to-noise ratios (S/N) were 3 and 10, respectively. As shown in Table 1, the LODs and the LOQs of the three TPM's BZD metabolites were within the range of 3.20-4.90 ng/g and 10.1-17.4 ng/g, respectively. The analysis recoveries and precisions were evaluated at three concentrations (25.0, 100, 500 ng/g) of three TPM's BZD metabolite standards spiked to blank celery cabbage. Table 2 showed that the average recoveries ranged from 82.6% to 113% and the precisions were within the range of 1.60%-18.3%, suggesting that this method exhibited good accuracy and reproducibility, and that it could be used for determining TPM's BZD metabolites in actual celery cabbage samples.
|
|
Table 1 Calibration curves of three TPM's known BZD metabolites spiked to celery cabbage. |
|
|
Table 2 Recoveries and precisions (Intra-and Inter-day) of the known BZD metabolites of TPM in celery cabbage. |
Using the proposed method, the TPM's BZD metabolite residues in positive celery cabbage samples were detected throughout the experimental period. The concentrations of BZ, 2-NH2BZ, and MBC in the samples were calculated by calibration curves. M-1, the newly discovered TPM's BZD metabolite was relatively quantified due to the lack of pure standard. Fig. 4 exhibited all quantitative results of TPM's BZD metabolites in celery cabbage samples. The average peak area ratio (analyte/internal standard) vs. time curves were plotted. The peak areas of all BZD compounds in celery cabbage reached maximum on day 4 after the spray with TPM and then decreased to be undetectable on day 10. Therefore, it could be concluded that celery cabbage sprayed with TPM within 10 days could not be consumed.

|
Download:
|
| Fig. 4. Curves of the peak areas of TPM's BZD metabolites in celery cabbage versus the collected time after spraying TPM onto celery cabbage. | |
In this study, a strategy of SiO2@NiO SPE combined with HPLC-PIS-MS and HR-MS/MS was used to identify and quantify TPM's BZD metabolites in celery cabbage. Four BZD metabolites were authors thank Xi-Zhou Hu (Hubei Academy of Agricultural identified and then monitored during the period after TPM was Sciences) for planting celery cabbage and spraying TPM. sprayed onto celery cabbage by using this strategy. Of these four BZD metabolites, one metabolite has not been reported previously. Comprehensive profiling of TPM's BZD metabolites in celery cabbage after TPM spraying was proposed. The results demon-strated the applicability of the method for monitoring TPM's metabolites residues in various vegetables.
AcknowledgmentsThe National Natural Science Foundation of China (No. 31671929, 21635006 and 31670373) supported this work. The authors thank Xi-Zhou Hu (Hubei Academy of Agricultural Sciences) for planting celery cabbage and spraying TPM.
Appendix A. Supplementary dataSupplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.cclet.2019.07.065.
| [1] |
A. Uclés, A. Valverde, M.D.G. García, et al., Anal. Methods 7 (2015) 9158-9165. DOI:10.1039/C5AY01048A |
| [2] |
Y. Xu, N. Song, Q. Zhang, et al., Food Chem. 221 (2017) 205-213. DOI:10.1016/j.foodchem.2016.10.009 |
| [3] |
Y. Soeda, S. Kosaka, T. Noguchi, Agric. Biol. Chem. 36 (1972) 931-936. DOI:10.1080/00021369.1972.10860358 |
| [4] |
C. Tejada-Casado, D. Moreno-González, F.J. Lara, et al., J. Chromatogr. A 1490 (2017) 212-219. DOI:10.1016/j.chroma.2017.02.023 |
| [5] |
J.P. Rouchaud, J.R. Decallonne, J.A. Meyer, Pestic. Sci. 8 (1977) 31-34. DOI:10.1002/ps.2780080105 |
| [6] |
D. Dalvie, E. Smith, A. Deese, et al., Drug Metab. Dispos. 34 (2006) 709-717. DOI:10.1124/dmd.105.008094 |
| [7] |
A. Nuñez, S.J. Lehotay, A.R. Lightfield, Rapid Commun. Mass Spectrom. 30 (2016) 813-822. DOI:10.1002/rcm.7508 |
| [8] |
L. Stuchlíková, R. Jirásko, L. Skálová, et al., Chemosphere 157 (2016) 10-17. DOI:10.1016/j.chemosphere.2016.05.015 |
| [9] |
O. Zamora, E.E. Paniagua, C. Cacho, et al., Anal. Bioanal. Chem. 393 (2009) 1745-1753. DOI:10.1007/s00216-009-2631-1 |
| [10] |
M. Majewsky, D. Castel, L.L. Dret, et al., J. Pharm. Biomed. Anal. 151 (2018) 151-158. DOI:10.1016/j.jpba.2017.12.056 |
| [11] |
A.C. Chukwudebe, P.G. Wislocki, D.R. Sanson, et al., J. Agric. Food Chem. 42 (1995) 2964-2969. |
| [12] |
D. Lugomer, M. Pavlicek, D. Bilandžić, et al., Mljekarstvo 67 (2017) 231-238. |
| [13] |
F. Zheng, H.M. Xiao, Q.F. Zhu, et al., Food Chem. 284 (2019) 279-286. DOI:10.1016/j.foodchem.2019.01.071 |
| [14] |
H. Sun, Q.W. Yu, H.B. He, et al., J. Agric. Food Chem. 64 (2015) 356-363. |
| [15] |
B. Palakshi Reddy, P. Iniyavan, S. Sarveswari, et al., Chin. Chem. Lett. 25 (2014) 1595-1600. DOI:10.1016/j.cclet.2014.06.026 |
| [16] |
I.S. Lee, N. Lee, J. Park, et al., J. Am. Chem. Soc. 128 (2006) 10658-10659. DOI:10.1021/ja063177n |
| [17] |
X.M. He, G.T. Zhu, W. Lu, et al., J. Chromatogr. A 1405 (2015) 188-192. DOI:10.1016/j.chroma.2015.05.040 |
| [18] |
Q.W. Yu, H. Sun, K. Wang, et al., Food Anal. Meth. 10 (2017) 2892-2901. DOI:10.1007/s12161-017-0837-y |
 2020, Vol. 31
2020, Vol. 31