b State Key Laboratory of Chemical Resource Engineering, Beijing University of Chemical Technology, Beijing 100029, China;
c CAS Key Laboratory of Standardization and Measurement for Nanotechnology, CAS Center for Excellence in Nanoscience, National Center for Nanoscience and Technology, Beijing 100190, China
In the past two decades, in response to cancer diagnosis and treatment, the number of new chemical entities (NCEs) and screenable drug targets has increased dramatically [1, 2]. However, 40% of preclinical and clinical drug candidates were eliminated due to metabolite toxicity [3]. Therefore, it is critically important to identify and verify the drug metabolites to predict drug safety in the early stages of drug discovery. Animal experiments are commonly used in in vitro drug toxicity studies. Due to inevitable heterogeneity, complex operations and a large amount of manpower and material resources requirements, it is not a high-throughput assessment method [4]. In addition, another in vitro models have widely been performed on plastic dishes, which are a two-dimensional platform that neglects the natural three-dimensional microenvironment. It may cause changes in cell phenotype and genetic information [5]. Hence, it is urgent to develop a three-dimensional culture tools in vitro. In order to better represent the in vivo cell biology, developing a simulation platform for co-culture of tumor cells and stromal cells can be used to explore the complex drug resistance during cancer treatment.
The rapid development of microfluidics has created many opportunities and possibilities for the study of biology and pharmacology [6, 7]. Instead of conventional methods to construct three-dimensional globule-culture models [8-11], microfluidic technology can achieve automated production based on the flexibility of fluid manipulation in space and time, mainly including micro-molding technique [12] and droplet technology [13]. In particular, droplet technology is a widely used method. Biological imitation matrices such as agar, fibrin, sodium alginate, polyethylene glycol can be introduced to support cell-matrix interactions, and it also reduces external shear stress to minimize cell damage. Microfluidic droplet technology can control the size of three-dimensional spheres well. It is a fast, reliable high-throughput biological research platform.
Besides accurately building physiologically relevant micro-environments with high spatial and temporal resolution, the microfluidic platform has powerful integration capabilities. Coupled with other analytical detectors [14, 15], cellular metabo-lism, and drug screening can be assessed. Especially, mass spectrometry (MS) as a highly sensitive, high-resolution structural analysis tool has shown tremendous analytical potential [16]. Many research reports have proposed to couple microfluidic chips with mass spectrometry to achieve simultaneous assessment of cytotoxicity and drug absorption, and the metabolic characteristics of many anticancer drugs have been analyzed, such as genistein [17], acetaminophen [18] and cyclophosphamide [19].
Here, we propose an integrated microfluidic platform to couple with mass spectrometry, which consists of three separate components to study the drug resistance due to the co-culture of tumor-endothelial cell: (1) establishment of a droplet micro-capsule model, (2) dynamic culture and drug stimulation, and (3) drug metabolism mass spectrometry analysis. The three-dimen-sional hydrogel sphere culture shows good biocompatibility, the inner sealed cells exhibit high activity. Based on the flexible design of the microfluidic chip, the compartmentalized microcapsules were prepared for co-culture of two heterogeneous cells. After pretreatment, the paclitaxel drug was extracted from metabolic medium and analyzed by electrospray ionization quadrupole time-of-flight mass spectrometry (ESI-Q-TOF MS). The difference of drug absorption between the co-culture system and the mono-culture system has been observed obviously. The integrated microfluidic device is a powerful tool to study cell metabolism and anticancer drug screening.
The integrated microfluidic platform includes two independent PDMA chips. As shown in Fig. 1, for the 1st chip, we design a droplet-templated chip to generate cell-laden hydrogel micro-capsules; and for 2nd chip, we culture and stimulate the purified microcapsules on-chip. Finally, we coupled ESI-Q-TOF MS detector to study drug absorption after pretreating the metabolic medium by solid phase extraction column (SPE).
|
Download:
|
| Fig. 1. Schematic illustration of integrated microfluidic device. Including three consecutive steps: (1) co-culture of tumor (A549 cells) and endothelial cells (HUVECs) based on droplet-templated alginate microcapsule, (2) dynamic perfusion culture and continuous drug stimulation, and (3) paclitaxel metabolites detection by ESI-Q-TOF MS. | |
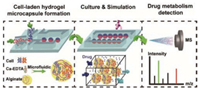
For generating cell-laden microcapsules, we employed alginate hydrogel as scaffold to provide 3D microenvironment for cells culture. Alginate mixing with Ca-EDTA solutions as disperse phase was introduced into inner microchannel, and HFE-7500 containing acid as continuous phase was injected into outer microchannel. At the joint, the disperse phase mixed with cells was squeezed into spherical microcapsule due to the interfacial tension between two immiscible solutions (Fig. 2A). H+ could diffuse into the interior of alginate, triggering Ca-EDTA to release Ca2+, and Ca2+ and alginate unit achieved gelation quickly in-situ. Through this microfluidic method, we get a mass of monodisperse microcapsules. After finished collection, we removed the oil solution from the bottom of tubes and added 50 μL fresh medium to resuspend microcapsules, then transferred into the micro-pit of the 2nd chip for subsequent culture (Fig. 2B). The structure of micro-pit could trap cell-laden microcapsules in-situ during dynamic culture medium perfusion. The solidified microcapsules could maintain stable structure during the working process (Fig. 2C).
|
Download:
|
| Fig. 2. (A) Schematic of 1st microfluidic chip for generating alginate microcapsules (a) and corresponding bright field image (b). (B) Schematic of 2nd microfluidic chip for cell-laden microcapsules culturing and stimulation. (C) The images of cell-laden microcapsules collected in oil (a) and cell culture medium (b), respectively. Scale bars are 100 and 50 μm, respectively. | |
Next, we evaluated the survival status of cells encapsulated in the microgel. We selected two kinds of cells (HUVEC and A549 cells) to be encapsulated into microcapsules respectively, and then collected into 2nd chip. Controlling 5 μL/h perfuse rate of medium for continuous culture, the cell kept high viability during a long period. As shown in Fig. 3, HUVEC cells proliferated during 7 days and A549 cells exhibited a tendency of aggregation inside the microcapsules.

|
Download:
|
| Fig. 3. Viability of encapsulated cells in the alginate microcapsules. The two kinds of cells (HUVEC and A549) were stained by the Calcein AM/EthD-1 kit. Scale bars are 100 μm. | |
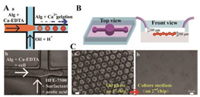
Based on the flexible design of microfluidic chip, we can get diversified structures of microcapsules. By designing droplet masks with different numbers of disperse phases (Fig. S1 in Supporting information), we produced microcapsules with up to three compartments as shown in Fig. 4A. The rapid Ca-alginate gelation ensured distinct partition interface. The mixed fluorescent polystyrene nanoparticles clearly displayed subareas of each microcapsules. We chose the two-compartment microcapsule to construct tumor-endothelial co-culture model. A549 cells, HUVEC cells were stained with red and green cell tracker reagent respectively, and were mixed with green and red fluorescent polystyrene nanoparticles-labeled two independent alginate phases separately. Then we produced corresponding co-culture microcapsule as Fig. 4B showed. Different cells were distributed in the separated compartment. The two-compartment hydrogel microsphere encapsulated heterogeneous cells in a micron-scale interval. It is convenient to contact with labor cells closely and material exchange. The microfluidic method allows a large amount of production.

|
Download:
|
| Fig. 4. (A) Confocal fluorescent images of multicompartment microcapsules with (a) one-, (b) two-, and (c) three-compartment microcapsules by designed microfluidic devices with different numbers of the disperse phase channels. (B) Schematic of encapsulating two kinds of heterogeneous cells in two-compartment microcapsules (a). The bright field images (b) and the confocal fluorescent images (c) of two-compartment microcapsules for encapsulating HUVEC cells and A549 cells inside. HUVEC, A549 were labelled with cell tracker Violet BMQC and Green CMFDA, respectively. Microcapsules are labelled with green and red fluorescent polystyrene nanomicrogels to distinguish the two compartments. (C) A parallel test of culturing cell-laden microcapsules on the 2nd chip. Scale bars are 50 μm. | |
For subsequent parallel testing, we stated that the micro-capsules were collected for 15 min under the same preparation condition. Then the collected microcapsules were transferred to the 2nd chip and connected to an external syringe pump for 24 h of continuous drug stimulation. As shown in Fig. 4C, tiny portable chips enable high throughput. The outlet of the each chip was connected with a tube to export the metabolic medium. This is a simple and convenient way to not only achieve on-chip culture and stimulation, but also allow the observation of cell states in-situ.
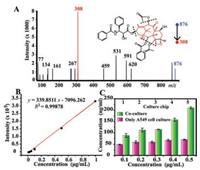
Furthermore, we detected the dose-dependent drug effect. Culture medium containing paclitaxel metabolites was pretreated by SPE columns. Firstly, we activated the columns using methanol and deionized water, then loaded the collected medium and followed by washing with water. Finally eluted by methanol and detected by ESI-Q-TOF-MS. The specific peaks of [PTX + Na]+ at m/z 876 and the representative fragment peaks at m/z 308 were clearly observed (Fig. 5A). The linearity of the chip-MS method was investigated with serial concentrations of PTX in medium ranging from 0.01 μg/mL to 1 μg/mL, with the linear equation of y = 339.8511x-7096.262. It displayed a satisfactory linear response (R2 = 0.9981) (Fig. 5B).
|
Download:
|
| Fig. 5. Detection of paclitaxel metabolites on the microfluidic chip-MS coupled platform. (A) The product ion scan spectra of 876 (m/z), the peak of fragment ion is at m/z 308. (B) The calibration curve of paclitaxel in culture medium. (C) The accumulation of paclitaxel in the culture medium of cells incubated with varied concentrations of paclitaxel from 0.1 μg/mL to 0.5 μg/mL for 24 h on-chip. | |
Moreover, we compared the difference of PTX absorption in co-cultured and mono-cultured A549 cells. Obviously, under the same concentration of drugs, the cells alone cultured showed a greater amount of drug absorption, while the experimental group co-cultured with endothelial cells absorbed less paclitaxel (Fig. 5C). Similar results have been reported in our previous reports [20]. It is further illustrated that co-cultured cells exhibit higher resistance to chemotherapeutic agents and are associated with more natural cell activity under co-culture conditions. These results indicate that the fast and sensitive drug absorption analysis platform can be used to partially evaluate clinical pharmacological effects.
In conclusion, we describe an integrated microfluidic chip platform to couple with MS detector to study drug metabolism based on a biological simulation model. We encapsulate A549 and HUVEC Cells into a two-compartment microcapsules for co-culture. Cells preserve high viability in the 3D, dynamic microenvironment. On-chip culture achieves continuous drug stimulation, and MS detection shows the significant difference of drug absorption between mono-culture and co-culture models. It is a high throughput, high sensitive method and has great potential in drug toxicity and drug screening fields.
AcknowledgmentThis work was supported by the National Natural Science Foundation of China (Nos. 21727814, 21435002 and 21621003).
Appendix A. SupplementSupplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.cclet.2019.07.036.
| [1] |
H.M. Geysen, F. Schoenen, D. Wagner, R. Wagner, Nat. Rev. Drug Discov. 2 (2003) 222-230. DOI:10.1038/nrd1035 |
| [2] |
E.F. Petricoin, K.C. Zoon, E.C. Kohn, J.C. Barrett, L.A. Liotta, Nat. Rev. Drug Discov. 1 (2002) 683-695. DOI:10.1038/nrd891 |
| [3] |
A.P. Li, Drug Discov. Today 6 (2001) 357-366. DOI:10.1016/S1359-6446(01)01712-3 |
| [4] |
T. Wang, Y. Deng, G.W. Qu, et al., Chin. Chem. Lett. 30 (2019) 1043-1050. DOI:10.1016/j.cclet.2019.01.011 |
| [5] |
G.Y. Lee, P.A. Kenny, E.H. Lee, M.J. Bissell, Nat. Methods 4 (2007) 359-365. DOI:10.1038/nmeth1015 |
| [6] |
T.Y. Yuan, D. Gao, S.F. Li, Y.Y. Jiang, Chin. Chem. Lett. 30 (2019) 331-336. DOI:10.1016/j.cclet.2018.07.013 |
| [7] |
C. Song, D. Gao, T.Y. Yuan, et al., Chin. Chem. Lett. 30 (2019) 1038-1042. DOI:10.1016/j.cclet.2019.02.017 |
| [8] |
H.L. Ma, Q. Jiang, S.Y. Han, et al., Mol. Imaging 11 (2012) 487-498. DOI:10.2310/7290.2012.00012 |
| [9] |
J.M. Kelm, N.E. Timmins, C.J. Brown, M. Fussenegger, L.K. Nielsen, Biotechnol. Bioeng. 83 (2003) 173-180. DOI:10.1002/bit.10655 |
| [10] |
J.B. Kim, Semin. Cancer Biol. 15 (2005) 365-377. DOI:10.1016/j.semcancer.2005.05.002 |
| [11] |
K. Ino, A. Ito, H. Honda, Biotechnol. Bioeng. 97 (2007) 1309-1317. DOI:10.1002/bit.21322 |
| [12] |
K. Kwapiszewska, A. Michalczuk, M. Rybka, R. Kwapiszewski, Z. Brzozka, Lab Chip 14 (2014) 2096-2104. DOI:10.1039/C4LC00291A |
| [13] |
C.G. Liu, W.C. Zheng, R.X. Xie, et al., Chin. Chem. Lett. 30 (2019) 457-460. DOI:10.1016/j.cclet.2018.09.010 |
| [14] |
O. Gefen, O. Fridman, I. Ronin, N.Q. Balaban, Proc. Natl. Acad. Sci. U. S. A. 111 (2014) 556-561. DOI:10.1073/pnas.1314114111 |
| [15] |
H.J. Song, J.M. Rosano, Y. Wang, et al., Lab Chip 15 (2015) 1320-1328. DOI:10.1039/C4LC01253D |
| [16] |
M.S. Jie, S.F. Mao, H.F. Li, J.M. Lin, Chin. Chem. Lett. 28 (2017) 1625-1630. DOI:10.1016/j.cclet.2017.05.024 |
| [17] |
Q.S. Chen, J. Wu, Y.D. Zhang, J.M. Lin, Anal. Chem. 84 (2012) 1695-1701. DOI:10.1021/ac300003k |
| [18] |
S.F. Mao, D. Gao, W. Liu, H.B. Wei, J.M. Lin, Lab Chip 12 (2012) 219-226. DOI:10.1039/C1LC20678H |
| [19] |
J. Wu, M.S. Jie, X.L. Dong, H.B. Qi, J.M. Lin, Rapid Commun. Mass Spectrom. 30 (2016) 80-86. DOI:10.1002/rcm.7643 |
| [20] |
L. Lin, X.X. Lin, L.Y. Lin, et al., Anal. Chem. 89 (2017) 10037-10044. DOI:10.1021/acs.analchem.7b02593 |
 2020, Vol. 31
2020, Vol. 31 

