b State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China
Pseudomonas aeruginosa (P. aeruginosa) is a critical human pathogen, which causes life-threatening infections and ranks first in the WHO list of priority pathogen research and development [1]. Due to the excessive intrinsic and acquired resistance mechanisms, multidrug-resistant (MDR) P. aeruginosa poses a serious threat to public health. In developed countries, P. aeruginosa is responsible for both community-and hospital-acquired infections, including hospital-acquired pneumonia (HAP), ventilator-associated pneumonia (VAP), and bloodstream infections (BSI) [2-4]. In addition, P. aeruginosa is a common pathogen in cystic fibrosis (CF) patients, where it most established chronic infection that accompanies the patient until the end of life [4]. The P. aeruginosa DK2 is an isolated strain from CF patients since 1973, its lineage persists in the lung environment for decades and could develop into an independent sub-system once it infected patients [5, 6]. The infections cause a significant proportion of morbidity and mortality in CF patients [7].
Polymyxin B (PB) is an ancient peptide antibiotic, which has been used in CF patients who infected with MDR Gram-negative bacteria during the past 30 years [8]. As one of the few available and effective antibiotics for chronic infected CF patients, PB is considered to be the last-line of defense against life-threatening infections caused by Gram-negative bacteria. However, with the increase use in clinical practice, P. aeruginosa developed resistance to PB recently [4, 8]. There are two strategies to respond the emergency of resistance: One is developing new antibiotics with novel modes of action against resistant bacteria; The other is finding synergists of PB to restore the susceptibility of resistant bacteria. In view, synergistic treatment could preserve the last class of antibiotics and reduce the emergency of drug resistance [9]. Repositioning of approved drugs is a good strategy to discover new synergists because this strategy is effective, quick and costeffective and there are some successful cases. For instance, the combination of aminoglycoside and penicillin is known to be used for treatment of Enterococcal infections, the combination of trimethoprim and sulfamethoxazole exhibit good efficacy against a number of pathogens [10]. Moreover, in 2018, Craig R. et al. discovered rifampicin in combination with colistin displayed growth-inhibition at levels below corresponding clinical breakpoints [9]. However, there is little research about synergists of PB.
In order to find good synergist of PB, we systematically performed a screen of 970 approved drugs synergized with PB against the clinical isolated MDR P. aeruginosa DK2 from Professor Lars Jelsbak of Technical University of Denmark [11], which is severely resistant to PB, MIC = 512 μg/mL. The primary screening was proceeded with PB at 32 μg/mL (1/16 of the minimal inhibitory concentration (MIC)) and the approved drugs at 100 μmol/L, which is an appropriate concentration enabled the selection of bioactive compounds, and the growth inhibition was monitored at 24 h after inoculation. The results showed that there were 131 active drugs with > 87% growth inhibition for the DK2, and about half of them were antibacterial agents. In this study, we focused on the effective antibacterial agents and the concentration was set at 50 μmol/L and 25 μmol/L for secondary screening. Finally, 19 fluoroquinolones which displayed potent adjuvant function with PB at 25 μmol/L were discovered (Fig. 1).
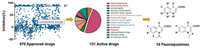
|
Download:
|
| Fig. 1. The screening process of fluoroquinolones synergy with PB. | |
To determine whether fluoroquinolones were additive or synergistic in the combinations with PB, the fractional inhibitory concentration (FIC) index was used as a measure of the interaction between two antimicrobial agents. FIC index of 1, ≤ 0.5 and ≥ 4 indicate no interaction, synergy and antagonism, respectively [12]. To achieve a wide range evaluation, we compared the interactions of fluoroquinolones at 1 μg/mL, which is a concentration not affected the growth rate and the bacterial numbers (Fig. S1 in Supporting information). As shown in Table 1, most tested fluoroquinolones displayed a strong synergy (FIC index ≤ 0.25). Especially, combined with gemifloxacin (GEM), sparfloxacin (SPA), enrofloxacin (ENR), ciprofloxacin (CIP), sarafloxacin (SAR) and moxifloxacin (MXF), PB exhibited high therapeutic potential with an obvious reduction in MIC below the clinical susceptible breakpoint (2 μg/mL) [13]. Encouragingly, 4096-fold reduction in MIC of PB (reduced 512 μg/mL to 0.125 μg/mL) was observed in combination with GEM. We further examined GEM in different concentrations combined with PB, the results showed that the synergistic effect was concentration-dependent, and the optimal synergistic concentration of GEM was 1 μg/mL (Table S1 in Supporting information).
|
|
Table 1 Combination studies of fluoroquinolones with PBa against the DK2. |
According to the results described above, GEM, SPA, ENR and CIP were identified as the potent synergists of PB for further experiment. In order to examine whether the synergy is effective in the class of peptide antibiotics, FIC index checkboard test was also carried on colistin (CL). CL is another peptide antibiotic which has been used in clinical practice. The main difference between the molecules of CL and PB is that the latter contains phenylalanine. Antimicrobial susceptibility test showed that the DK2 was more resistant against CL (MIC > 1024 μg/mL) than PB. We tested the ability of CL against the DK2 combined with four fluoroquinolones (GEM, SPA, ENR, CIP). As shown in Table 2, the synergistic effect with CL could also be observed, while the MIC of CL failed to reduce below the susceptible breakpoint (2 μg/mL) in combination with fluoroquinolones.
|
|
Table 2 Combination studies of GEM, SPA, ENR and CIP with CLa against the DK2. |
In order to investigate whether fluoroquinolones potentiation is conserved beyond the clinical isolated DK2 strains, the timekilling kinetic assay of optimal compound GEM was conducted on two wild-type P. aeruginosa strains (PAO1, MPAO1). Before the start, MIC of GEM and PB for each wild-type strain had been tested (Table S2 in Supporting information) and we fixed GEM at 0.25 × MIC (0.25 μg/mL) in the assay, which is a concentration not significantly affected the growth of bacteria, and the concentration of PB was determined by the results of growthcurve in PAO1 and MPAO1 (Figs. S2A and S2C in Supporting information) In the time-killing kinetic assay, synergy was defined as a reduction ≥ 2 log CFU/mL with combination respect to the active single agent [14]. In the case of PAO1, 0.25 μg/mL GEM with 0.25 μg/mL PB significantly declined the bacterial cell count by 3.7 log CFU/mL (> 2 log CFU/mL) compared with PB alone at 8 h (Fig. S2B in Supporting information). However, as for MPAO1, the combination of 0.25 μg/mL GEM with 0.25 μg/mL or 0.5 μg/mL PB was consistent with the growth inhibition of 0.25 μg/mL or 0.5 μg/mL PB alone and no synergy was observed (Fig. S2D in Supporting information). Consequently, the combination of GEM and PB showed the best synergistic effect against the clinical isolated strain DK2, the weak synergistic effect in PAO1 within 8 h, but no synergy in MPAO1.
In order to investigate the synergy mechanism, we explored the influence of GEM on outer membrane permeability via measuring the uptake of the hydrophobic fluorophore N-phenyl-1-naphthylamine (NPN) by bacteria after addition of GEM, with 1% TritonX-100 and 10 mmol/L EDTA as positive control [15]. The results validated that the synergistic concentrations of GEM could increase the permeability of all tested P. aeruginosa strains cell membrane (Fig. 2 and Fig. S3 in Supporting information). Especially, the permeability of cell membrane displayed significant concentration-dependent in DK2, and cell membrane uptake capacity of 1 μg/mL GEM was even stronger than the positive control 10 mmol/L EDTA (Fig. 2). This phenomenon was not discovered in other strains (PAO1 and MPAO1). The result supported the optimal synergistic effect of GEM and PB in DK2.

|
Download:
|
| Fig. 2. Permeabilization of outer membrane by GEM in DK2 cells. TritonX-100(1%) and EDTA (10 mmol/L) were used as positive control. | |
In summary, through comprehensive screening and experiment, fluoroquinolones were chosen as peptide antibiotic synergists for inhibiting the growth of P. aeruginosa DK2. Encouragingly, GEM showed the highest synergy with PB, leading a 4096-fold MIC reduction at 1 μg/mL. In addition, SPA, ENR, CIP, SAR and MXF also achieved an obvious reduction in MIC of PB below the clinical susceptible breakpoint (2 μg/mL). Time-killing kinetic assay displayed that the combination of GEM and PB behaved the best synergistic effect in the clinical isolated strain DK2, the weak synergistic effect in PAO1 within 8 h, and no synergy in MPAO1. NPN uptake assay demonstrated that GEM greatly enhanced the permeability of cell outer membranes in the P. aeruginosa DK2, which might explain its synergy mechanism. Therefore, fluoroquinolones represent attractive synergists to address the emerging threat of polymyxin-resistant infections.
AcknowledgmentsThis work was supported by the National Key R&D Program of China (No. 2017YFB0202600), the National Natural Science Foundation of China (Nos. 21672064, 21702061, 81861138047), the Innovative Research Team of High-level Local Universities in Shanghai, the National Special Fund for State Key Laboratory of Bioreactor Engineering (No. 2060204), "Shu Guang" project supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (No. 14SG28) and the Shanghai Sailing Program (No. 17YF1403600).
Appendix A. Supplementary dataSupplementary material related to this article can befound, in the online version, at doi:https://doi.org/10.1016/j.cclet.2019.07.063.
| [1] |
E. Tacconelli, N. Magrini, World Health Organization, Geneva, 2017. https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25FebET_NM_WHO.pdf.
|
| [2] |
F. Imperi, F. Massai, C.R. Pillai, et al., Antimicrob. Agents Chemother. 57 (2013) 996-1005. DOI:10.1128/AAC.01952-12 |
| [3] |
L. Puzniak, D.D. DePestel, A. Srinivasan, et al., Antimicrob. Agents Chemother. 63 (2019) e02564-18. DOI:10.1128/AAC.02564-18 |
| [4] |
S. Nagarajan, P. Shanmugavelan, M. Sathishkumar, et al., Chin. Chem. Lett. 25 (2014) 419-422. DOI:10.1016/j.cclet.2013.12.017 |
| [5] |
R.L. Marvig, M.S.R. Sondergaard, S. Damkiaer, et al., Antimicrob. Agents Chemother. 56 (2012) 4519-4521. DOI:10.1128/AAC.00630-12 |
| [6] |
L. Yang, L. Jelsbak, R.L. Marvig, et al., Proc. Natl. Acad. Sci. U. S. A. 108 (2011) 7481-7486. DOI:10.1073/pnas.1018249108 |
| [7] |
A. Folkesson, L. Jelsbak, L. Yang, et al., Nat. Rev. Microbiol. 10 (2012) 841-851. DOI:10.1038/nrmicro2907 |
| [8] |
M. Zhu, P. Liu, Z.W. Niu, Chin. Chem. Lett. 28 (2017) 703-708. DOI:10.1016/j.cclet.2016.10.001 |
| [9] |
C.R. Macnair, J.M. Stokes, L.A. Carfrae, et al., Nat. Commun. 9 (2018) 458-465. DOI:10.1038/s41467-018-02875-z |
| [10] |
G.D. Wright, Trends Microbiol. 24 (2016) 862-871. DOI:10.1016/j.tim.2016.06.009 |
| [11] |
M.H. Rau, R.L. Marvig, G.D. Ehrlich, et al., Environ. Microbiol. 14 (2012) 2200-2211. DOI:10.1111/j.1462-2920.2012.02795.x |
| [12] |
L. Ejim, M.A. Farha, S.B. Falconer, et al., Nat. Chem. Biol. 7 (2011) 348-350. DOI:10.1038/nchembio.559 |
| [13] |
C.L.S. Institute, M100-S27, 44, 2017, ISBN 1-56238-1-56238-805-3.
|
| [14] |
R. Ayerbe-Algaba, M.L. Gil-Marqués, M.E. Jiménez-Mejías, et al., Front. Cell. Infect. Microbiol. 8 (2018) 348. DOI:10.3389/fcimb.2018.00348 |
| [15] |
X. Yang, S. Goswami, B.K. Gorityala, et al., J. Med. Chem. 60 (2017) 3913-3932. DOI:10.1021/acs.jmedchem.7b00156 |
 2020, Vol. 31
2020, Vol. 31 



