b School of Pharmacy, Second Military Medical University, Shanghai 200433, China;
c Key Laboratory of Marine Drugs, Ministry of Education, School of Medicine and Pharmacy, Ocean University of China, Qingdao 266003, China
Protein glycosylation is one of the most ubiquitous and complex PTMs found in all domains of life and plays a pivotal role in regulating the biological complexity of eukaryotes [1-3]. With the advancement of proteomics, an increasing number of novel protein glycosylation modifications have been gradually discovered and recognized [4-6]. However, the N-glycosylation of Arg residue in proteins were only reported in very rare cases [7] until Shao Feng's [8] and Hartland's [9] groups discovered and illuminated an unprecedented N-acetylglucosamine (GlcNAc) transferase derived from EPEC type-Ⅲ-secreted effector NleB, which specifically modified conserved arginine on several human proteins bearing a death domain, including TRADD, FAS-associated death domain protein (FADD), TNF receptor 1 (TNFR1) and the kinase RIPK1. Surprisingly, it was shown that arginine N-GlcNAcylation could functionally block the assembly of a DISC, effectively preventing EPEC-infected cells from undergoing apoptosis and necroptosis. The particular glycosylated modification revealed a new model by which bacteria counteract host defenses, and also a potential antimicrobialtarget.
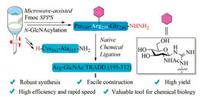
To monitor other important arginine N-GlcNAcylated modification of proteins in human proteomics, we have reported the first synthesis of Arg-GlcNAc peptides via a silver-promoted solid phase glycosylationprocess and obtained the first antibodies which can specifically recognize Arg-GlcNAc modification in a sequence-independent manner [10]. Even though we have made some progress about Arg-GlcNAc modification, the discovery of Arg-GlcNAc glycoprotein and its related research on biological functions need to be further studied. For example, there are several Arg in the vicinity of Arg235 of TRADD death domain, does the Arg-GlcNAc modification of these sites affect the self-polymerization and physiological function of TRADD death domain? Hence, synthesis of Arg-GlcNAc TRADD (195–312) (Scheme 1) or its congeners thereafter would facilitate detailed studies on this significant filed.
|
Download:
|
| Scheme 1. The synthetic strategy toward Arg-GlcNAc TRADD (195–312). | |
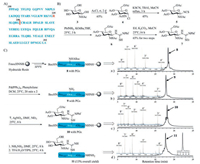
Structurally, the primary protein structure of Arg-GlcNAc TRADD (195–312) comprises 117 amino acid residues (Fig. 1A) without intramolecular disulfide bonds, N-GlcNAcylated site is at Arg235. To undertake this synthesis, three synthetic methodology technologies were crucial to our endeavors, including microwave-assisted SPPS, silver-promoted on-resin guanidinylation and hydrazide-based NCL. Microwave-assisted SPPS provides a highly efficient, economical and time-saving strategy for preparing longish peptidyl fragments (even over 50 amino acids), in addition, it avoids the usage of prohibitively expensive ChemMatrix resin, pseudoproline and isoacyl dipeptides [11, 12]. Silver-promoted on-resin guanidinylation serves as an alternative method for the introduction of the Arg-GlcNAc moiety and it showed good compatibility on long peptides unlike glycosyl-amino acid building blocks [13]. On the basis of seminal work of Kent and co-workers [14], NCL has become one of the most important methods to chemoselectively link a pair of unprotected peptide and protein segments, one functionalized as a C-terminal thioester and another with an N-terminal cysteine (Cys) residue to generate proteins [15]. In order to expand its concept, considerable effort has been devoted to tuning the C-terminal acyl donor thioesters used in NCL [16, 17]. Hydrazide-based NCL developed by Liu group has proved versatile and effective, and its utility exemplified in the chemical synthesis of many complex proteins [18-23].
|
Download:
|
| Fig. 1. (A) The sequence of Arg-GlcNAc TRADD (195–312). (B) Synthesis of building block N-glycosyl-S-ethyl-isothiourea 7. (C) Solid-phase synthetic route, analytical HPLC chromatogram (λ = 214 nm) and HR-Q-TOF-MS spectrum of glycopeptide 11. | |
For programmatic expediency, we carried out the forward synthesis first by preparing the key building block N-glycosyl-S-ethyl-isothiourea 7 according to previous work (Fig. 1B) [10, 24]. In brief, glycosyl chloride 4 was prepared starting from 2-acetamino-2-deoxy-d-glucose 3 by acetyl chloride. Then 4 was treated with potassium thiocyanate (KSCN) and tetrabutylammonium iodide (TBAI) in anhydrous acetonitrile to yield glycosyl isothiocyanate 5 in moderate yield. The glycosyl Pbf-thiourea 6 was synthesized by pre-mixing Pbf-NH2 with tBuOK, and immediately alkylated with iodoethane to afford the N-glycosyl-S-ethyl-isothiourea 7 in 65% yield over the two steps.
Given there is only one cysteine in Arg-GlcNAc TRADD (195–312), we envisioned that the it might be audaciously assembled from just two peptidyl fragments, Pro195-Gly240 11 (45 residues) and Cys241-Ala312 12 (72 residues), respectively. Fragment 8 (with protecting groups (PGs)) was prepared by a microwave peptide automatic synthesizer (CEM Liberty Lite) with Fmoc-hydrazine resin as the solid support, and Fmoc-Orn (Alloc)−OH was installed at the glycosylated site (Alloc, allyloxycarbonyl) under standard coupling (DIC (N, N'-diisopropylcarbodiimide)/Oxyma pure) and Fmoc-deprotection conditions (25% piperidine in dimethylformamide (DMF) with 0.1 mol/L Oxyma pure). Then, the Alloc group was removed using Pd(PPh3)4 (tetrakis (triphenylphosphine) palladium) and phenylsilane to yield fragment 9 (with PGs) on the resin. The free amino side chain of 9 (with PGs) subsequently underwent guanidinylation with AgNO3 and 7 to afford glycopeptide 10 (with PGs) on the resin. Next, the acetyl groups on GlcNAc were removed with 5% NH2NH2 in DMF. These conditions facilitated complete reaction and highly purity as judged by analytical test cleavage with cocktail trifluoroacetic acid (TFA) (TFA/Triisopropylsilane (TIPS)/H2O = 95/2.5/2.5, v/v/v) followed by high performance liquid chromatography (HPLC) and high resolution-quadrupole-time of flight-mass MS (HR-Q-TOF-MS) analysis (Fig. 1C). From here, the resin-bound glycopeptide was treated with above cocktail TFA to release glycopeptide 11, the crude product 11 was purified by semi-preparative reverse-phase HPLC to afford the desired glycopeptide 11 in an isolated yield of 13% as calculated from the resin loading.
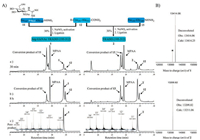
On the other side, ultralong fragment 12 also was successfully synthesized, purifed, and lyophilized in just 4 days under the help of microwave synthesizer (with 98% purity). With key segments 11 and 12 in hand, we attempted to assemble Arg-GlcNAc TRADD (195–312) by hydrazide-based NCL (Fig. 2A). Firstly, the peptide 11 was dissolved in ligation buffer (6 mol/L guanidinium chloride, 200 mmol/L NaH2PO4, pH 3.0) pre-cooled in an ice bath (−10 ℃). Then NaNO2 was added dropwise and the reaction was incubated for 30 min at −10 ℃ to convent the hydrazide to the acyl azide. Next 4-mercaptophenylacetic acid (MPAA) was added and the pH was adjusted to 5.0 for 5 min to generate the thioesterpeptide. Finally, the N-terminal Cys peptide 12 was added and the pH value of the ligation solution was adjusted to 6.5–6.7 to initiate the NCL reaction. Presumably due to the hydrophobicity of 12 and longish length for two segments, the ligation between 11 and 12 was incomplete even after 8 h. The targeted crude glycoprotein 1 was purified by semi-preparative reverse-phase HPLC in an isolated yield of 30%. Applying the same strategy, we also smoothly obtained TRADD (195–312) 2 without Arg-GlcNAc modification as a control, all the highly pure samples were verifed by HR-Q-TOF-MS (Fig. 2B).
|
Download:
|
| Fig. 2. (A) Synthetic route, analytical HPLC chromatogram (λ = 214 nm) and HR-Q-TOF-MS spectrum of Arg-GlcNAc TRADD (195–312) 1 and wild-type TRADD (195–312) 2. (B) Deconvoluted HR-Q-TOF-MS spectra of 1 and 2 (*denotes the loss of N-acetylglucosamine during ionization in mass spectrometer). | |
The difference in secondary structure between 1 and 2 was further compared by circular dichroism (CD) spectrum in PBS buffer at a final concentration of 50 μmol/L [25, 26]. As show in Fig. 3A, the CD spectrum of 1 was remarkably consistent with wild-type protein 2, which indicates Arg-GlcNAc modification did not induce conformational change of 2. In addition to CD, Fourier transform infrared (FTIR) spectroscopy also evidenced their extremely similar geometric construction (Fig. 3B). These result implied, besides glycosylation, other unknown conditions might be the cause for the self-polymerization of TRADD death domain.

|
Download:
|
| Fig. 3. (A) CD spectra of 1 (in blue) and 2 (in green). (B) FTIR characterization of 1(in blue) and 2 (in green). | |
In summary, we have successfully synthesized TRADD death domain with arginine N-GlcNAcylation and corresponding wild-type TRADD death domain on milligram scale by combining microwave-assisted SPPS, silver-promoted on-resin guanidinylation and peptidyl hydrazide-based NCL. Considering that Arg-GlcNAc modification is a novel PTM and potential antimicrobial target, our chemical synthetic method allows functional glycosylation into peptide at atom level precision like other N-linked homogeneous glycoproteins, which will be of valuable tool for chemical biology [27], and provides a representative paradigm for the rapid synthesis of other proteins with Arg-GlcNAc modification.
AcknowledgmentsWe are grateful to the National Natural Science Foundation of China (Nos. 91849129, 21807112), PLA Youth Medical Science and Technology Youth Development Program (No. 16QNP086) and Foundation of Second Military Medical University (No. 2016JS11).
Appendix A. Supplementary dataSupplementary material related to this article canbe found, in the online version, at doi:https://doi.org/10.1016/j.cclet.2019.05.010.
| [1] |
K. Ohtsubo, J.D. Marth, Cell 126 (2006) 855-867. DOI:10.1016/j.cell.2006.08.019 |
| [2] |
D.J. Gill, H. Clausen, F. Bard, Trends. Cell. Biol. 21 (2011) 149-158. DOI:10.1016/j.tcb.2010.11.004 |
| [3] |
K.W. Moremen, M. Tiemeyer, A.V. Nairn, Nat. Rev. Mol. Cell. Biol. 13 (2012) 448-462. |
| [4] |
J. Lassak, E.C. Keilhauer, M. Furst, et al., Nat. Chem. Biol. 11 (2015) 266-270. DOI:10.1038/nchembio.1751 |
| [5] |
J. Qiu, M.J. Sheedlo, K. Yu, et al., Nature 533 (2016) 120-124. DOI:10.1038/nature17657 |
| [6] |
P. Zhou, Y. She, N. Dong, et al., Nature 561 (2018) 122-126. DOI:10.1038/s41586-018-0433-3 |
| [7] |
D.G. Singh, J. Lomako, W.M. Lomako, et al., FEBS Lett. 376 (1995) 61-64. DOI:10.1016/0014-5793(95)01247-6 |
| [8] |
S. Li, L. Zhang, Q. Yao, et al., Nature 501 (2013) 242-246. DOI:10.1038/nature12436 |
| [9] |
J.S. Pearson, C. Giogha, S.Y. Ong, et al., Nature 501 (2013) 247-251. DOI:10.1038/nature12524 |
| [10] |
M. Pan, S. Li, X. Li, et al., Angew. Chem. Int. Ed. 53 (2014) 14517-14521. DOI:10.1002/anie.201407824 |
| [11] |
Q. Qu, M. Pan, S. Gao, et al., Adv. Sci. 5 (2018) 1800234. DOI:10.1002/advs.201800234 |
| [12] |
M. Pan, S. Gao, Y. Zheng, et al., J. Am. Chem. Soc. 138 (2016) 7429-7435. DOI:10.1021/jacs.6b04031 |
| [13] |
X. Li, R. Krafczyk, J. Macosek, et al., Chem. Sci. 7 (2016) 6995-7001. DOI:10.1039/C6SC02889F |
| [14] |
P.E. Dawson, T.W. Muir, I. Clark-Lewis, et al., Science 266 (1994) 776-779. DOI:10.1126/science.7973629 |
| [15] |
A.C. Conibear, E.E. Watson, R.J. Payne, et al., Chem. Soc. Rev. 47 (2018) 9046-9068. DOI:10.1039/C8CS00573G |
| [16] |
S.S. Kulkarni, J. Sayers, B. Premdjee, et al., Nat. Rev. Chem. 2 (2018) 0122. DOI:10.1038/s41570-018-0122 |
| [17] |
B. Yan, W. Shi, L. Ye, et al., Curr. Opin. Chem. Biol. 46 (2018) 33-40. DOI:10.1016/j.cbpa.2018.03.018 |
| [18] |
G.M. Fang, J.X. Wang, L. Liu, et al., Angew. Chem. Int. Ed. 51 (2012) 10347-10350. DOI:10.1002/anie.201203843 |
| [19] |
J.S. Zheng, S. Tang, Y.K. Qi, et al., Nat. Protoc. 8 (2013) 2483-2495. DOI:10.1038/nprot.2013.152 |
| [20] |
Y.C. Huang, G.M. Fang, L. Liu, et al., Natl. Sci. Rev. 3 (2016) 107-116. DOI:10.1093/nsr/nwv072 |
| [21] |
G.M. Fang, Y.M. Li, F. Shen, et al., Angew. Chem. Int. Ed. 50 (2011) 7645-7649. DOI:10.1002/anie.201100996 |
| [22] |
Z.M. Wang, W.L. Xu, L. Liu, et al., Nat. Chem. 8 (2016) 698-704. DOI:10.1038/nchem.2517 |
| [23] |
J.S. Zheng, M. Yu, Y.K. Qi, et al., J. Am. Chem. Soc. 136 (2014) 3695-3704. DOI:10.1021/ja500222u |
| [24] |
S. Wang, L. Corcilius, P.P. Sharp, et al., Bioorg. Med. Chem. 25 (2017) 2895-2900. DOI:10.1016/j.bmc.2017.03.012 |
| [25] |
Y. Wu, Y.H. Li, X. Li, et al., Chem. Sci. 8 (2017) 7368-7373. DOI:10.1039/C7SC02420G |
| [26] |
X. Li, W.D. Tolbert, H.G. Hu, et al., Chem. Sci. 10 (2019) 1522-1530. DOI:10.1039/C8SC03275K |
| [27] |
U. Carlo, K. Yasuhiro, Curr. Opin. Chem. Biol. 46 (2018) 130-137. |
 2020, Vol. 31
2020, Vol. 31 

