In the past several decades, cancer has become one of the major diseases threatening human health. Among the many cancer treatments, chemotherapy is still the most commonly used means of treating cancer [1-3]. Moreover, people have made considerable efforts to develop therapeutic agents, such as doxorubicin (DOX) [4-6], camptothecin (CPT) [7, 8], docetaxel (DTX) [9, 10] and their analogues, etc. DOX is an anthracyline drug, which has shown substantial treatment potential to several cancers. The mecha-nisms of DOX causing cancer cells death is to damage topoisomer-ase-II-mediated DNA repair and generate free radicals [11]. Unsatisfactorily, free DOX is highly toxic to normal cells and tissues, furthermore, poor water solubility and easily cleared by reticuloendothelial system, which greatly limits the therapeutic effect of DOX [12-15].
In order to overcome this dilemma, drug delivery systems have been developed, in which polymer-drug conjugates is a relatively promising mean of treating cancer [16-18]. Polymeric prodrug is a form of drug covalently linked to polymer that is inactive during blood circulation and is activated when it reaches the site of the lesion [19, 20]. Amphiphilic polymeric prodrug especially, has aroused great attention due to the increased solubility of free insoluble drugs in biological fluid, prolonged circulation time, and that, under the protection of the polymer shell, the chemotherapy toxicity of small molecule drug is reduced and the absorption of drug by tumor tissues is enhanced by the enhanced permeability and retention (EPR) effect [21-24].
In recent years, stimuli-responsive polymeric drug delivery systems, which are responsive to the change of environmental stimuli for controlled drug release, have made significant progress [25-27]. These environmental stimuli include physical condition such as temperature, light, magnetic field, irradiation, ultrasound and chemical condition such as redox potential, enzymes, ionic strength, pH profile [28-30]. As an important signal, the pH values in tumors tissues (pH 6.2–6.9) and intracellular compartments, for instance, the endosomes and lysosomes (pH 4.5–6.0) are much lower than normal tissues and blood (pH 7.4) [31-33]. Therefore, based on the difference pH profiles, pH-responsive polymeric prodrug carriers can be designed. Many synthetic or modified polymers with good biocompatibility have been used in drug delivery systems, such as hyaluronic acid (HA) [34, 35], poly(ethylene glycol) (PEG) [36-39], poly(D, L-lactic-co-glycolic acid) (PLGA) [40, 41], poly(ε-caprolactone) (PCL) [42, 43], polyphosphoesters (PPEs) [44, 45], polypeptide [46-48], and the like. In particular, FDA-approved PEG and its analogues have excellent water solubility, which are widely applied as drug delivery carriers.
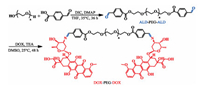
Although many PEG-based polymer prodrugs have been studied, the modification processes of most PEGylated prodrugs are cumbersome [49-51]. Herein, we design and synthesize a PEG-based pH-sensitive polymeric prodrug DOX-PEG-DOX, which has simple structure and easy to be prepared via Schiff-base reaction, as shown in Scheme 1. The materials, detailed synthesis processes, structural characterization methods and data are provided in the Supporting information (Figs. S1–S6 in Supporting information). For convenience of description, we used P1k, P2k and P4k to represent the prodrugs of DOX-PEG1k-DOX, DOX-PEG2k-DOX and DOX-PEG4k-DOX, respectively.
|
Download:
|
| Scheme 1. Synthesis routes of DOX-PEG-DOX polymeric prodrug. | |
The DOX loading capacity of prodrug nanoparticles was determined by UV–vis measurement after stirring in 1 mol/L HCl for 8 h. A calibration curve was made using DOX/HCl solutions with different DOX concentrations. The DOX content was calculated according to Eq. (1):

|
(1) |
where CDOX represents the concentration of DOX measured by UV–vis, CProdrug is the concentration of prodrug nanoparticles.
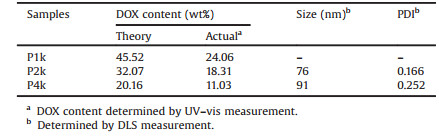
The results are listed in Table 1, from which one can find that the drug contents of P1k, P2k and P4k are 24.06%, 18.31% and 11.03%, respectively. The three prodrug nanoparticles have relatively high DOX contents compared to the conventional method to structure a polymeric drug delivery system. In order to obtain the prodrug nanoparticles with relatively uniform morphology and good stability, we choose P2k to further study.
|
|
Table 1 Characteristics of P1k, P2k, P4k nanoparticles. |
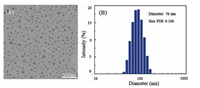
The morphologies of P2k prodrug nanoparticles were observed by TEM measurement, and the average particle diameters (
|
Download:
|
| Fig. 1. (A) TEM image of P2k nanoparticles and (B) the particle size distribution histogram corresponding to the TEM sample in PB 7.4 (scale bar 200 nm). | |
For investigating the pH responsiveness of P2k prodrug, the size change of the nanoparticles under the condition of pH 5.0 and 7.4 were measured by DLS, TEM and 1H NMR. As shown in Fig. 2, we can find that the average size of the nanoparticles has no obvious changes after 48 h under the condition of pH 7.4, indicating that the good stability of the prodrug nanoparticles. However, the size of P2k prodrug nanoparticles increased significantly and the particle size distribution had become wide with the increasing incubation time in the presence of pH 5.0. Moreover, we used TEM to study the morphology changes of P2k prodrug nanoparticles at pH 7.4 and pH 5.0 after 48 h, which was consistent with the DLS results as shown in Fig. S7 (Supporting information). This is due to that the structure of P2k prodrug nanoparticles were dissociated under acidic condition and the aggregation of PEG segment formed large particles. In addition, the pH sensitivity of Schiff-base bonds was proved by the characteristic chemical shift of the aldehyde groups at 10.0 ppm under pH 5.0 after 8 h, as shown in Fig. S8 (Supporting information).

|
Download:
|
| Fig. 2. Size change of P2k prodrug nanoparticles under different conditions of (A) pH 7.4 and (B) pH 5.0, as determined by DLS measurement. | |
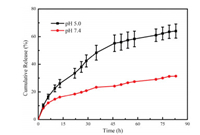
The cumulative DOX release behavior of acid-responsive P2k was studied at physiological pH 7.4 and intracellular acidic pH 5.0 at 37 ℃. As illustrated in Fig. 3, there are approximately 64% of DOX was released from P2k prodrug nanoparticles after incuba-tion for 84 h under the condition of pH 5.0, whereas the release of DOX was only about 31% under the condition of pH 7.4. It can be seen that the pH-responsive P2k prodrug nanoparticles contain-ing Schiff-base bonds could be cleaved under acidic condition, resulting in dissociation of nanoparticles to accelerate the release of DOX. Furthermore, P2k prodrug nanoparticles maintain relative stability in the in vivo environment, preventing premature DOX release.
|
Download:
|
| Fig. 3. In vitro DOX release curves for P2k prodrug nanoparticles at pH 7.4 and pH 5.0. The nanoparticles concentration was 1 mg/mL. Data were presented as mean standard deviation (n = 3). | |
According to our design, P2k and P4k nanoparticles can endocytose into cells and release the parent DOX in an acidic environment to inhibit the proliferation of tumor cells. Here, the ability of P2k and P4k nanoparticles to inhibit tumor cell proliferation was studied using MTT assays against HeLa cells and HepG2 cells with different DOX concentrations (0.019 mg/L to 9.50 mg/L). As illustrated in Fig. 4A, all the free DOX, P2k and P4k nanoparticles showed significant toxicity to HeLa cells, in which the best effect of inhibiting the proliferation of tumor cells is free DOX and the cytotoxicity of P4k is the lowest. Corresponding the half-maximal inhibitory concentration (IC50) values of free DOX, P2k, and P4k nanoparticles were determined to be 0.16 mg/L, 1.31 mg/L, and 1.61 mg/L, respectively. It is probably because of the rapid diffusion of free DOX through the cellular membrane and notably reducing the cells viability. Furthermore, the slow DOX release from P2k and P4k also caused the decrease of the cytotoxicity. On the other hand, high molecular weight PEG hindered the release of DOX from prodrug nanoparticles [49, 51]. The result of Fig. 4B is similar to that of Fig. 4A. The above results indicate that P2k and P4k have a good inhibitory effect on these both tumor cells.

|
Download:
|
| Fig. 4. In vitro cytotoxicity of free DOX, P2k and P4k with different DOX concentrations against (A) HeLa cells and (B) HepG2 cells after 72 h incubation. Data were presented as mean standard deviation (n = 4). | |
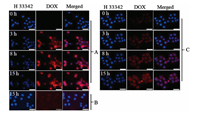
To visually verify whether prodrug nanoparticles can efficiently enter tumor cells, the cellular uptake behavior of P2k and P4k nanoparticles was observed against HeLa cells by fluorescence microscopy of the live cell imaging system. The cells were performed in Dulbecco's Modified Eagle medium with the same DOX concentration of P2k and P4k, and photos were taken at different time intervals to observe the endocytic process, while free DOX as a control group. As illustrated in Fig. 5A, the DOX red fluorescence of P2k was observed in HeLa cells after 3 h, and the red color gradually deepened with the extension of time. In addition, the red fluorescence of DOX was evident in the nucleus after 15 h incubation compared to the control group in Fig. 5B. This might be the result of the way of free DOX cellular internalization by diffusion, and could be easily pumped out of tumor cells [52]. In Fig. 5C, P4k can also internalization into HeLa cells quickly, but the fluorescence intensity is lower than that of P2k, it could be that molecular length of PEG impede the endocytosis process [49].
|
Download:
|
| Fig. 5. Live cell imaging system images of HeLa cells incubated with (A) P2k nanoparticles, (B) free DOX and (C) P4k nanoparticles for different times. The DOX dosage was 7.6 mg/L. For each panel, images from left to right show cell nuclei stained by H 33342 (blue), DOX fluorescence in cells (red), and overlays of the blue and red images. The scale bars corresponded to 50 mm in all images. | |
In summary, we used a facile way to prepare polymeric prodrugs based on different molecular weights of PEG by Schiff base reaction, which have good hydrophilicity, high drug loading and acid sensitivity. These prodrugs can self-assemble to form nanoparticles of uniform particle size (76 nm for P2k, 91 nm for P4k) with PEG as the shell and conjugated DOX as the core. P2k nanoparticles are stable under physiological conditions, and yet an accumulative DOX release of P2k nanoparticles was up to 64% at pH 5.0. In vitro cytotoxicity assay showed that P2k and P4k prodrug nanoparticles could effectively inhibit the proliferation of HeLa cells with IC50 of 1.31 mg/L and 1.61 mg/L, respectively, after 15 h of incubation. Take advantage of cellular uptake assays, we visually observed the endocytic process of prodrugs and confirmed that P2k and P4k nanoparticles could entered into HeLa cells to release DOX. Therefore, this pH-responsive PEGylated polymeric prodrug has promising potential in the application for cancer treatment.
AcknowledgmentsWe gratefully acknowledge the financial supports from the National Natural Science Foundation of China (No. 21374066), the Major Program of the Natural Science Project of Jiangsu Higher Education Institutions (No. 15KJA150007), Natural Science Foun-dation of Jiangsu Province (No. BK20171212), the Project Funded by the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions, and Soochow-Waterloo University Joint Project for Nanotechnology from Suzhou Industrial Park.
Appendix A. Supplementary dataSupplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.cclet.2019.04.052.
| [1] |
A. Ediriwickrema, W.M. Saltzman, ACS Biomater. Sci. Eng. 1 (2015) 64-78. DOI:10.1021/ab500084g |
| [2] |
W.P. Fan, B. Yung, P. Huang, X.Y. Chen, Chem. Rev. 117 (2017) 13566-13638. DOI:10.1021/acs.chemrev.7b00258 |
| [3] |
Z.R. Lu, P. Qiao, Mol. Pharm. 15 (2018) 3603-3616. DOI:10.1021/acs.molpharmaceut.8b00037 |
| [4] |
P. Menna, E. Salvatorelli, G. Minotti, Chem. Res. Toxicol. 21 (2008) 978-989. DOI:10.1021/tx800002r |
| [5] |
M. Cagel, E. Grotz, E. Bernabeu, et al., Drug Discov. Today 22 (2017) 270-281. DOI:10.1016/j.drudis.2016.11.005 |
| [6] |
C. Pe'rez-Arnaiz, N. Busto, J.M. Leal, B. García, J. Phys. Chem. B 118 (2014) 1288-1295. DOI:10.1021/jp411429g |
| [7] |
D.P. Curran, H. Liu, J. Am. Chem. Soc. 114 (1992) 5863-5864. DOI:10.1021/ja00040a060 |
| [8] |
K. Li, J.J. Ou, S.H. Gao, Angew. Chem. Int. Ed. 55 (2016) 14778-14783. DOI:10.1002/anie.201607832 |
| [9] |
J. Dubois, Le Goff M.T., F.G. Voegelein, et al., Biorg. Med. Chem. 3 (1995) 1357-1368. DOI:10.1016/0968-0896(95)00115-W |
| [10] |
C. Lucatelli, F. Viton, Y. Gimbert, A.E. Greene, J. Org. Chem. 67 (2002) 9468-9470. DOI:10.1021/jo026460n |
| [11] |
C.F. Thorn, C. Oshiro, S. Marsh, et al., Pharmacogenet. Genom. 21 (2011) 440-446. DOI:10.1097/FPC.0b013e32833ffb56 |
| [12] |
A.P. Blum, J.K. Kammeyer, A.M. Rush, et al., J. Am. Chem. Soc. 137 (2015) 2140-2154. DOI:10.1021/ja510147n |
| [13] |
P.W. Burridge, Y.F. Li, E. Matsa, et al., Nat. Med. 22 (2016) 547-556. DOI:10.1038/nm.4087 |
| [14] |
P. Ganguly, A. Breen, S.C. Pillai, ACS Biomater. Sci. Eng. 4 (2018) 2237-2275. DOI:10.1021/acsbiomaterials.8b00068 |
| [15] |
C.L. Zhu, Y.N. Xia, Chem. Soc. Rev. 46 (2017) 7668-7682. DOI:10.1039/C7CS00492C |
| [16] |
B. Louage, L. Nuhn, M.D. Risseeuw, et al., Angew. Chem. Int. Ed. 55 (2016) 11791-11796. DOI:10.1002/anie.201605892 |
| [17] |
G.Q. Ma, J. Liu, J.L. He, M.Z. Zhang, P.H. Ni, ACS Biomater. Sci. Eng. 4 (2018) 2443-2452. DOI:10.1021/acsbiomaterials.8b00429 |
| [18] |
J.J. Sun, Y.H. Liu, Y.C. Chen, et al., J. Control. Release 258 (2017) 43-55. DOI:10.1016/j.jconrel.2017.05.006 |
| [19] |
J. Khandare, T. Minko, Prog. Polym. Sci. 31 (2006) 359-397. DOI:10.1016/j.progpolymsci.2005.09.004 |
| [20] |
N. Larson, H. Ghandehari, Chem. Mater. 24 (2012) 840-853. DOI:10.1021/cm2031569 |
| [21] |
F. Greco, M.J. Vicent, Adv. Drug Deliv. Rev. 61 (2009) 1203-1213. DOI:10.1016/j.addr.2009.05.006 |
| [22] |
D. Li, X.R. Feng, L. Chen, et al., ACS Biomater. Sci. Eng. 4 (2018) 539-546. DOI:10.1021/acsbiomaterials.7b00856 |
| [23] |
H. Maeda, Bioconjug. Chem. 21 (2010) 797-802. DOI:10.1021/bc100070g |
| [24] |
Y.L. Zhai, X. Zhou, L.N. Jia, et al., Polymers 9 (2017) 698. DOI:10.3390/polym9120698 |
| [25] |
N. Kamaly, B. Yameen, J. Wu, O.C. Farokhzad, Chem. Rev. 116 (2016) 2602-2663. DOI:10.1021/acs.chemrev.5b00346 |
| [26] |
J. Li, D.J. Mooney, Nat. Rev. Mater. 1 (2016) 16071. DOI:10.1038/natrevmats.2016.71 |
| [27] |
K. Zhang, P.P. Yang, J.P. Zhang, L. Wang, H. Wang, Chin. Chem. Lett. 28 (2017) 1808-1816. DOI:10.1016/j.cclet.2017.07.001 |
| [28] |
X. Pang, Y. Jiang, Q.C. Xiao, et al., J. Control. Release 222 (2016) 116-129. DOI:10.1016/j.jconrel.2015.12.024 |
| [29] |
F. Seidi, R. Jenjob, D. Crespy, Chem. Rev. 118 (2018) 3965-4036. DOI:10.1021/acs.chemrev.8b00006 |
| [30] |
J.H. Xia, T.Y. Li, C.J. Lu, H.P. Xu, Macromolecules 51 (2018) 7435-7455. DOI:10.1021/acs.macromol.8b01597 |
| [31] |
S. Bazban-Shotorbani, M.M. Hasani-Sadrabadi, A. Karkhaneh, et al., J. Control. Release 253 (2017) 46-63. DOI:10.1016/j.jconrel.2017.02.021 |
| [32] |
F.H. Meng, Z.Y. Zhong, J. Feijen, Biomacromolecules 10 (2009) 197-209. DOI:10.1021/bm801127d |
| [33] |
S.J. Sonawane, R.S. Kalhapure, T. Govender, Eur. J. Pharm. Sci. 99 (2017) 45-65. DOI:10.1016/j.ejps.2016.12.011 |
| [34] |
Z.X. Gu, X.X. Wang, R. Cheng, et al., Acta Biomater. 80 (2018) 288-295. DOI:10.1016/j.actbio.2018.09.022 |
| [35] |
Y. Zhang, K.Q. Wu, H.L. Sun, et al., ACS Appl. Mater. Interfaces 10 (2018) 1597-1604. DOI:10.1021/acsami.7b17718 |
| [36] |
S.S. Banerjee, N. Aher, R. Patil, J. Khandare, J. Drug Deliv. 2012 (2012) 103973. |
| [37] |
R. Reid, M. Sgobba, B. Raveh, et al., Macromolecules 48 (2015) 7359-7369. DOI:10.1021/acs.macromol.5b01598 |
| [38] |
W.G. Xu, J.X. Ding, X.S. Chen, Biomacromolecules 18 (2017) 3291-3301. DOI:10.1021/acs.biomac.7b00950 |
| [39] |
J.C. Chen, J.Z. Li, J.H. Liu, et al., Chin. Chem. Lett. 26 (2015) 1319-1321. DOI:10.1016/j.cclet.2015.05.050 |
| [40] |
C.J. Bowerman, J.D. Byrne, K.S. Chu, et al., Nano Lett. 17 (2017) 242-248. DOI:10.1021/acs.nanolett.6b03971 |
| [41] |
B. Sivakumar, R.G. Aswathy, R.R. Aburto, et al., Biomater. Sci. 5 (2017) 432-443. DOI:10.1039/C6BM00621C |
| [42] |
J. Hu, M.Z. Zhang, J.L. He, P.H. Ni, RSC Adv. 6 (2016) 40858-40868. DOI:10.1039/C6RA07420K |
| [43] |
C. Wei, Y. Zhang, H. Xu, et al., J. Mater. Chem. B 4 (2016) 5059-5067. DOI:10.1039/C6TB01040G |
| [44] |
X.Q. Du, Y. Sun, M.Z. Zhang, et al., ACS Appl. Mater. Interfaces 9 (2017) 13939-13949. DOI:10.1021/acsami.7b02281 |
| [45] |
H. Elzeny, F.W. Zhang, E.N. Ali, et al., Drug Des. Dev. Ther. 11 (2017) 483-496. DOI:10.2147/DDDT.S128503 |
| [46] |
S.Q. Yang, L.Z. Zhou, Y. Su, et al., Chin. Chem. Lett. 30 (2019) 187-191. DOI:10.1016/j.cclet.2018.02.015 |
| [47] |
T.W. Wang, C.W. Yeh, C.H. Kuan, et al., Acta Biomater. 58 (2017) 54-66. DOI:10.1016/j.actbio.2017.06.008 |
| [48] |
Y.H. Zheng, Y.L. Cheng, J.J. Chen, et al., ACS Appl. Mater. Interfaces 9 (2017) 3487-3496. DOI:10.1021/acsami.6b15245 |
| [49] |
W.H. Lin, L. Yin, T.T. Sun, et al., Biomacromolecules 19 (2018) 1625-1634. DOI:10.1021/acs.biomac.8b00083 |
| [50] |
Y.C. Tao, S.W. Liu, Y. Zhang, et al., Polym. Chem. 9 (2018) 878-884. DOI:10.1039/C7PY02108A |
| [51] |
H.R. Wang, J.L. He, D.L. Cao, et al., Polym. Chem. 6 (2015) 4809-4818. DOI:10.1039/C5PY00569H |
| [52] |
Y. Yu, C.K. Chen, W.C. Law, et al., Polym. Chem. 6 (2015) 953-961. DOI:10.1039/C4PY01194E |
 2019, Vol. 30
2019, Vol. 30