b College of Life Sciences, Fujian Normal University, Fuzhou 350117, China;
c Key Laboratory for Analytical Science of Food Safety and Biology (MOE & Fujian Province), Department of Chemistry, Fuzhou University, Fuzhou 350116, China
A portable, sensitive and specific method for detecting and quantifying disease-related biomarkers, especially at a lowconcentration level, is very important for evaluating the therapy and determining recurrence or metastasis [1, 2]. A pH meter, a scientific device for measuring the hydrogen-ion activity in waterbased solutions, works by measuring the difference in electrical potential between known reference electrode and the measuring pH electrode [3]. Potential of pH electrode depends on the logarithm of the concentration (or more precisely activity) of hydronium ion on the basis of the Nernst equation [4]. Thanks to portability and easy operation, different detection protocols and strategies based on pH meter have been developed in the fields of analytical and bioanalytical chemistry, e.g., for caner biomarkers [5, 6], telomerase activity [7], thrombin inhibitor drugs [8], and cancer cells [9]. In these systems, the pH change of the detection solutions usually derived from the catalytic reaction of bioactive enzymes (e.g., glucose oxidase, GOx, toward the glucose substrate to produce gluconic acid) or classical chemical reactions (e.g., the reaction between CO32- and H+). Typically, GOx is one of the most widely used alternatives since it is easily labeled onto the biomolecules (e.g., antibodies, nanomaterials or oligonucleotides) [10, 11]. Kinetics and equilibrium of the virtually every reaction occurring in the detection solution depends on the solution pH value [12].
For the successful development of pH meter-based detection systems, the signal amplification is very crucial for detection of low-abundance biomarkers. Undoubtedly, enzyme-based signalamplification strategies are widely used in this respect, e.g., enzyme-linked immunosorbent assay (ELISA), because a singlemolecular enzyme usually causes the conversion of numerous enzymatic substrates per min [13, 14]. Unfortunately, the labeled enzymatic number on one biomolecule is limited because of the steric hinerance effect (e.g., one antibody can be usually labeled with one enzyme molecule) [15]. In contrast, the emergence of nanobiotechnology opens a new horizon for the signal amplification since nanomaterials have a high surface coverage and good biocompatibility [16-20]. Recent reports have been focused on the signal amplification by combining enzyme labels with nano labels in the sensing systems [21, 22]. Relative to enzyme labels alone, introduction of nano labels can indeed increase the labeled amount of enzyme molecules to some extent. Unfavorably, a monolayer of enzyme molecules can be often conjugated to the surface of nanostructures, thus controlling the finite biomolecules due to the limitation of surface coverage [23]. An alternative strategy that is exploring three-dimensional space of the nanomaterials by coupling with molecular biological amplification technologies (e.g., rolling circle amplification, RCA, and hybridization chain reaction, HCR) and enzyme labels would be advantageous [24-27]. HCR, a toehold-mediated strand-displacement reaction, has attracted great interest because of its simple protocol and excellent amplification efficiency [28]. During this process, the initiator strand can readily trigger the cross-opening of two DNA hairpins to yield nicked double-helix similar with the alternating copolymer [29]. To this end, our motivation of this study is to construct threedimensional nano labeling system with the enzymatic amplification for the fabrication of electrochemical immunoassay.
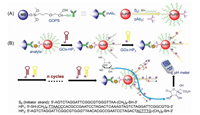
Herein, we reports on the proof-of-concept of pH meter-based potentiometric immunoassay for portable and powerful detection of disease-related biomarkers (squamous cell carcinoma antigen, SCCA, used as a model in this case because it is an importance tumor marker in patients with the uterine cervical squamous cell carcinoma) (Scheme 1). To construct such a nanoparticle-based sensing system, monoclonal mouse anti-human SCCA capture antibody (mAb1) is initially conjugated covalently onto magnetic bead (MB) to form mAb1-MB-based immunosensing probe, whereas polyclonal rabbit anti-human detection antibody (pAb2) and initiator strand (S0) are modified onto gold nanoparticle (AuNP) to form the pAb2-AuNP-S0 recognition element. The detailed process can be found in Supporting information. This system mainly consists of mAb1-MB, pAb2-AuNP-S0 and two GOxlabeled hairpins (HP1 and HP2). In the presence of target SCCA, a sandwiched immunoreaction is carried out between mAb1-MB and pAb2-AuNP-S0 to generate a high-ratio immunocomplex between S0 and the analyte. Upon GOx-labeled hairpins introduction, all the conjugated initiator strands on the AuNP undertake an unbiased strand-displacement reaction and open two alternating hairpin DNA probes in sequence, thereby resulting in the formation of three-dimensional long nicked double-helix network around the gold nanoparticle. In this case, numerous GOx molecules are concatenated together by the double-stranded DNA and encapsulated into the network. Upon addition of enzyme substrate (glucose), each of GOx molecules oxidizes glucose into gluconic acid and hydrogen peroxide, thus causing the pH shift of the detection solution, which can be readout on a handheld pH meter. With the increasing target SCCA, the large-amount gluconic acid molecules are generated thanks to introduction of many pAb2-AuNP-S0 conjugates. By monitoring the pH change, we can exactly evaluate the concentration of target SCCA in the sample. Therefore, the whole detection process is relatively simple and low cost for the detection of cancer biomarkers.
|
Download:
|
| Scheme 1. .Schematic illustration of pH meter-based magneto-controlled immune-HCR assay for detection of squamous cell carcinoma antigen (SCCA) on mAb1 capture antibody-coated magnetic bead (MB) by using initiator S0 strand/pAb2 secondary antibody-labeled gold nanoparticle (AuNP) as the detection antibody, accompanying initiator strand-triggered hybridization chain reaction (HCR) between two glucose oxidase (GOx)-conjugated hairpins (HP1 and HP2). (A) Preparation processes of mAb1-MB and pAb2-AuNP-S0. (B) Immune-HCR reaction and pH meter measurement. | |
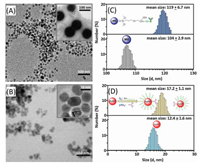
As mentioned above, the assay mainly includes immunoreaction, HCR reaction and the oxidization of glucose. To realize our design, one important precondition is to prepare the mAb1-MB and pAb2-AuNP-S0 conjugates for the progression of the sandwiched immunoreaction. Figs. 1A and B give typical transmission electron microscope (TEM; H-7650, Hitachi Instruments, Tokyo, Japan) images of mAb1-MB and pAb2-AuNP-S0, respectively. Vaguely, a layer of bright brim could be also observed around the nanoparticles (Fig. 1A and B, insets), which derived from the labeled DNA or antibodies. Since the labeled DNA/antibodies were a kind of biomolecules or proteins, they might be carbonized at the high potential. Moreover, the mean sizes were 119 · 6.7 nm for mAb1-MB (Fig. 1C, top) and 17.2 ·1.1 nm for pAb2-AuNP-S0 (Fig. 1D, top), obtained from dynamic light scattering (DLS; Zetasizer Nano S90, Malvern, London, UK) data. Obviously, the sizes of the corresponding nanostructures after modification with the biomolecules were more than those before conjugation (top vs. bottom, Figs. 1C and D). The increased size stemmed from the labeled biomolecules. In addition, we also employed Fourier transform infrared spectroscopy (FTIR), zeta potential and UV-vis absorption spectroscopy to characterize the conjugates, respectively (Fig. S1 for mAb1-MB and Fig. S2 for pAb2-AuNP-S0 in Supporting information). These results revealed the successful synthesis of mAb1-MB and pAb2-AuNP-S0 conjugates by our designed route.
|
Download:
|
| Fig. 1. TEM images of (A) mAb1-MB and (B) pAb2-AuNP-S0 (insets: magnification images); (C, D) DLS data of (C) magnetic beads before (bottom) and after (top) conjugation with mAb1 antibody, and (D) gold nanoparticles before (bottom) and after (top) labeling with initiator strand (S0) and pAb2 antibody. | |
Except for the immunoreaction, the initiator strand (S0)-triggered HCR reaction is also very crucial for the signal amplification. To demonstrate this issue, we utilized gel electrophoresis to investigate HP1 and HP2 in the absence and presence of the initiator strand. Experimental results indicated that mixture of HP1 with HP2 alone did not cause their self-hybridization reaction, and introduction of the initiator strand could trigger the HCR reaction between HP1 and HP2, resulting in formation of long nicked double-helices (Fig. S3 in Supporting information). Next, we preliminarily evaluated the feasibility of pH meter-based immunoassay by coupling with HCR reaction by using 1.0 ng/mL SCCA as an example. The pH values were collected and registered on a handheld pH meter after each step in the presence of 2.0 mol/L glucose in PBS (10 mmol/L, pH 7.5; optimized because GOx exhibited high catalytic efficiency at this pH) (Fig. 2A). Relative to the background signal (column a), the pH values of the detection solutions were not almost changed toward mAb1-MB (column b), SCCA + mAb1-MB (column c) and S0-AuNP-pAb2 + SCCA + mAb1-MB (column d). Upon progression of HCR reaction between two GOxlabeled hairpins by the carried S0 on the AuNP, significantly, an obvious shift in the pH value (column e) was acquired relative to column a. In contrast, the absence of target SCCA in this system did not cause the significant change in the pH, indicating that pAb2-AuNP-S0 could not nonspecifically adsorb to the mAb1-MB. In this regard, our strategy can be utilized to detect target SCCA with pH meter readout.

|
Download:
|
| Fig. 2. (A) The pH readouts of different components in 10 mmol/L pH 7.5 PBS: (a) glucose, (b) mAb1-MB + glucose, (c) mAb1-MB + SCCA + glucose, (d) mAb1-MB + SCCA + pAb2-AuNP-S0 + glucose and (e) mAb1-MB + SCCA + pAb2-AuNP-S0 + GOx-HP1/GOx-HP2 + glucose; (B) pH responses of magneto-controlled immunoassay by using different signal amplification strategies: (a) GOx-pAb2, (b) GOx-AuNP-pAb2 and (c) S0-AuNP-pAb2 + GOx-HP1/GOx-HP2 (1.0 ng/mL SCCA used in these cases). | |
Logically, one question arises as to whether the developed immune-HCR assay can readily amplify the detectable signal. For comparison, another two labeling protocols including GOx-labeled pAb2 (GOx-pAb2) and GOx/pAb2-labled AuNP (GOx-AuNP-pAb2) were also used for detection of target SCCA with the same assay format (1.0 ng/mL used in this case). As seen from columns b and c in Fig. 2B, the immunoassays of using GOx-pAb2 and GOx-AuNP-pAb2 as the recognition elements exhibited low pH shifts relative to the background signal (column a). In contrast, a high pH change was achieved in this system by coupling pAb2-AuNP-S0 with the HCR reaction (column d). We might roughly estimate that one solid AuNP with 10 nm in diameter could conjugate 16 GOx molecules at most (note: The calculation is based on the spherical surface area [SNP = 4πrNP2] divided by the area of the GOx's radius-based circle [SGOx = πrB2], where rNP and rB stand for the AuNP radius and GOx radius [~ 5.0 nm in diameter], respectively). Compared with threedimensional HCR-based network on the AuNP, the immune-HCR assay could accommodate more GOx molecules in this system for the catalytic oxidation of glucose into gluconic acid, thus resulting in the signal amplification.
To achieve a high sensitivity, the following parameters were optimized, such as molar ratio for S0:pAb2 for preparation of S0-AuNP-pAb2 (5:2), immunoreaction time (30 min) and HCR reaction time (150 min) (Fig. S4 in Supporting information). Under optimum conditions, we studied the analytical properties of the immune-HCR assay toward SCCA standards. The shift in the pH value increased with the increasing SCCA concentration in the sample, and a good linear relationship between DpH and the logarithm of target SCCA concentration was acquired in the range of 0.01–10 ng/mL (Fig. 3A). The detection limit (LOD) was 5.7 pg/mL, as calculated at the 3sblank criterion (n = 13). Each data point represents the average value obtained from three different measurements. The maximum relative standard deviation (RSD) was 6.4%. Furthermore, we investigated the batch-to-batch precision and reproducibility of mAb1-MB and pAb2-AuNP-S0 toward three high-middle-low SCCA levels including 0.1, 1.0 and 10 ng/mL, and the RSD values were 11.7%, 9.2% and 12.4% (n = 3), respectively. These results indicated good reproducibility and precision of immune-HCR assay. To further highlight the merits of immune-HCR assay, the analytical properties including the linear range and LOD were compared with other portable SCCA detection schemes. As indicated from Table S1 (Supporting information), the LOD of our strategy was comparable with these methods.

|
Download:
|
| Fig. 3. (A) The pH responses (vs. background signal) of immune-HCR assay toward different-concentration SCCA standards. (B) The specificity of immune-HCR assay against 1.0 ng/mL SCCA, 100 ng/mL CEA, 100 ng/mL AFP, 100 ng/mL PSA, 100 U/mL CA 125 and 100 U/mL CA 15-3. | |
Next, we also monitored the specificity of immune-HCR assay against the non-targets, e.g., cancer antigen 15-3 (CA 15-3), carcinoembryonic antigen (CEA), cancer antigen 125 (CA 125), prostate-specific antigen (PSA) and alpha-fetoprotein (AFP). As shown in Fig. 3B, these non-targets including CEA, CA 125, PSA, AFP and CA 15-3 only caused relatively low signal shifts in comparison with the blank sample, whilst a significant pH change was achieved in the presence of only target SCCA, suggesting good specificity of immune-HCR assay. Furthermore, the storage stability of mAb1-MB and pAb2-AuNP-S0 was measured over a six-month period. When storing them at 4 ℃ and measuring intermittently (every 3-5 days) for 1.0 ng/mL SCCA (used as an example), they retained 97.3%, 92.7% and 90.8% (n = 3) of the initial signal at the 2nd, 4th and 6th month, respectively, suggesting an acceptable stability.
Finally, the accuracy of immune-HCR assay was investigated in comparison with the gold method (e.g., commercial human SCCA ELISA kit; Abcam) for human serum samples. Initially, we collected several human serum specimens including target SCCA from our hospital according to the rules of the local ethical committee (note: Informed consent was obtained from any experimentation with human subjects). Thereafter, these samples were determined by using the immune-HCR assay and ELISA kit, respectively. Table S2 (Supporting information) gives the results obtained from two methods. Evaluation of the accuracy between two methods was executed by using a classical t-test. Clearly, all the texp values in all cases were below tcrit (tcrit[2, 0.05] = 4.30), indicating good accuracy between two methods.
In conclusion, this work successfully demonstrated the development of an advanced immune-HCR assay with pH meter readout by coupling with enzyme-conjugated HCR reaction and nano labeling strategy. Functionalized magnetic beads and gold nano labels were used for construction of magneto-controlled immunosensing probes. Target-induced enzyme-conjugated HCR reaction was utilized for the signal amplification. Highlight of this study is adequately combining enzyme labels and nano labels with molecular biological amplification technique for the fabrication of nanoparticle-based portable immunoassay. Such a signal-amplified immunoassay is favorable for quantitative detection of the low-abundance proteins in disease diagnosis. Importantly, nanoparticles-based immunosensing probes can be suitable for use in mass production of magneto-controlled flow-through devices and open new opportunities for protein diagnostics and biosecurity.
AcknowledgmentsAuthors acknowledged financial support from the National Natural Science Foundation of China (No. 21675029) and the Health-Education Joint Research Project of Fujian Province (No. WKJ2016-2-15).
Appendix A. Supplementary dataSupplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.cclet.2019.03.045.
| [1] |
G. Sciutto, M. Zangheri, L. Anfossi, et al., Angew. Chem. Int. Ed. 57 (2018) 7385-7389. DOI:10.1002/anie.201713298 |
| [2] |
Z. Yu, Y. Tang, G. Cai, et al., Anal. Chem. 91 (2019) 1222-1226. DOI:10.1021/acs.analchem.8b04635 |
| [3] |
A. McLean, Acad. Psychiatry 41 (2017) 769.
|
| [4] |
Y. Zhang, J. Yang, J. Nie, et al., Chem. Commun. 52 (2016) 3474-3477. DOI:10.1039/C5CC09852A |
| [5] |
J. Liang, J. Wang, L. Zhang, et al., New J. Chem. 43 (2019) 1372-1379. DOI:10.1039/C8NJ05637D |
| [6] |
Y. Jiang, Z. Su, J. Zhang, et al., Analyst 143 (2018) 5271-5277. DOI:10.1039/C8AN01436A |
| [7] |
L. Wang, C. Chen, H. Huang, et al., Biosens. Bioelectron. 121 (2018) 153-158. DOI:10.1016/j.bios.2018.08.069 |
| [8] |
J. Wang, M. Song, C. Hu, et al., Anal. Chem. 90 (2018) 9366-9373. DOI:10.1021/acs.analchem.8b01979 |
| [9] |
X. Luo, H. Yang, H. Wang, et al., Anal. Chem. 90 (2018) 5803-5809. DOI:10.1021/acs.analchem.8b00218 |
| [10] |
R. Ren, G. Cai, Z. Yu, et al., Sens. Actuators B 265 (2018) 174-181. DOI:10.1016/j.snb.2018.03.049 |
| [11] |
A. Singh, S. Park, H. Yang, Anal. Chem. 85 (2013) 4863-4868. DOI:10.1021/ac400573j |
| [12] |
L. Miao, Z. Zhang, L. Jiao, et al., Chin. Chem. Lett. 28 (2017) 1878-1880. DOI:10.1016/j.cclet.2017.04.018 |
| [13] |
Y. Xianyu, J. Wu, Y. Chen, et al., Angew. Chem. Int. Ed. 57 (2018) 7503-7507. DOI:10.1002/anie.201801815 |
| [14] |
B. Zhang, D. Tang, I. Goryacheva, et al., Chem.-Eur. J. 19 (2013) 2496-2503. DOI:10.1002/chem.201203131 |
| [15] |
X. Pei, B. Zhang, J. Tang, et al., Anal. Chim. Acta 758 (2013) 1-18. DOI:10.1016/j.aca.2012.10.060 |
| [16] |
L. Luo, L. Zhang, R. Zeng, et al., Anal. Chem. 90 (2018) 9568-9575. DOI:10.1021/acs.analchem.8b02421 |
| [17] |
T. Song, W. Wang, L. Meng, et al., Chin. Chem. Lett. 28 (2017) 226-230. DOI:10.1016/j.cclet.2016.07.021 |
| [18] |
R. Ren, G. Cai, Z. Yu, et al., Anal. Chem. 90 (2018) 11099-11105. DOI:10.1021/acs.analchem.8b03538 |
| [19] |
M. Li, J. Chen, J. Pan, et al., Chin. Chem. Lett. 30 (2019) 541-544. DOI:10.1016/j.cclet.2018.11.017 |
| [20] |
J. Chen, Y. Huang, X. Yang, et al., Anal. Chim. Acta 1023 (2018) 89-95. DOI:10.1016/j.aca.2018.02.082 |
| [21] |
P. Zhu, Y. Zhang, S. Xu, et al., Chin. Chem. Lett. 30 (2019) 58-62. DOI:10.1016/j.cclet.2018.02.003 |
| [22] |
J. Shu, D. Tang, Chem.-Asian J. 12 (2017) 2780-2789. DOI:10.1002/asia.201701229 |
| [23] |
S. Lv, K. Zhang, Y. Zeng, et al., Anal. Chem. 90 (2018) 7086-7093. DOI:10.1021/acs.analchem.8b01825 |
| [24] |
R. Zeng, Z. Luo, L. Su, et al., Anal. Chem. 91 (2019) 2447-2454. DOI:10.1021/acs.analchem.8b05265 |
| [25] |
Z. Zhang, S. Lv, Z. Lin, et al., Biosens. Bioelectron. 101 (2018) 159-166. DOI:10.1016/j.bios.2017.10.031 |
| [26] |
Z. Qiu, J. Shu, D. Tang, Anal. Chem. 90 (2018) 1021-1028. DOI:10.1021/acs.analchem.7b04479 |
| [27] |
J. Chen, H. Qiu, M. Zhang, et al., Biosens. Bioelectron. 68 (2015) 550-555. DOI:10.1016/j.bios.2015.01.054 |
| [28] |
R. Lin, Q. Feng, P. Li, et al., Nat. Methods 15 (2018) 275-278. DOI:10.1038/nmeth.4611 |
| [29] |
S. Bi, S. Yue, S. Zhang, Chem. Soc. Rev. 46 (2017) 4281-4298. DOI:10.1039/C7CS00055C |
 2019, Vol. 30
2019, Vol. 30 

