b Britton Chance Center for Biomedical Photonics, Wuhan National Laboratory for Optoelectronics Huazhong University of Science and Technology, Wuhan 430074, China;
c MoE Key Laboratory for Biomedical Photonics, Collaborative Innovation Center for Biomedical Engineering, School of Engineering Sciences, Huazhong University of Science and Technology, Wuhan 430074, China
Hyperspectral stimulated Raman scattering (SRS) microscopy has been applied in biochemical field as an alternatively superior imaging technique without fluorescent label. It uses scattered light that generated by vibrations of chemical bonds in biomolecules instead [1-4]. The vibrational signals are specific to chemical bond, and thus provide a structure fingerprint by which molecules can be identified [3, 5, 6]. Therefore, Raman imaging could avoid the drawbacks, such as the intense interference of cells or tissues' auto fluorescence, poor photo-stability and photo-bleaching effect, which are inevitable in fluorescence imaging technique.
There are intrinsic Raman signals of biomolecules within cells, and the wavenumber of vibrational signals of chemical bond in these biomolecules locates at either lower than 1800 cm-1 or higher than 2800 cm-1. In order to minimize these background signals, exogenous Raman probes or modified endogenous biomolecules whose signals appear in the Raman-silent region of cells (1800-2800 cm-1) have been explored as an alternative strategy for imaging [1, 7]. Research along this orientation has developed two methods for recognition of specific small molecules in vivo. One is isotopically modifying biomolecules, the other is modifying biomolecules with triple bond (alkyne, nitrile, azide) [8-10]. Such as isotope or triple bond modified deoxyribonucleoside, amino acid, glucose and lipid have been reported and incorporated into cells to test their metabolic process via Raman imaging [11-16]. Among these approaches, alkyne or polyyne modified molecules show stronger Raman signal in the silent region of cell, free from the disturbing signals of endogenous biomolecules.
Among all the cellular organelles, lysosomes are the terminal one in the endocytic pathway [17]. They are intracellular membrane-bound organelles with a diameter of about 500 nm and contain more than 60 degradative enzymes, which enable cell to break down various biomolecules, including peptides, nucleic acids, lipids, or even the invading virus particles and bacteria [18, 19]. In addition, some decomposed components were reused as available nutrients to the cell [19]. As a result, the versatile capacity for degradation of lysosomes plays a fundamental and crucial role in cell maintenance. On the contrary, the dysfunction of lysosomes will abate its ability to degrade endogenous biomolecules and invading compounds, and reduce lysosomal activity will further lead to an increase in vulnerability and viral infectivity for cell [20, 21]. Hence, it is of great value to monitor lysosomes and explore its cellular function. Fluorescent-based methods employing lysosomes-targeting probes have been widely investigated [22-25]. All these probes contain a secondary amine that is suited for the purpose because of their high sensitivity and specificity to the acidic microenvironment of lysosomes [26]. Consequently, we envisioned that Raman-based approach could also be exploited as novel high-performance probes for the visualization of lysosome when it was coupled with a secondary amine structure.
First, we surveyed the structure-Raman shift and intensity of different alkynes. Among these various alkynyl structures, bisarylbutadiynes (BADY) has reported to be a good building block for generating strong Raman intensity, producing signal peaks about 25 times stronger than that of 5-ethynyl-20- deoxyuridine (EdU) on average (Fig. 1) [7, 27]. Therefore, we envisioned its potentiality to be used as a sensitive and specific Raman tag for visualizing cellular organelles such as lysosome after conjugating with a suitable targeting group. Here, we describe the synthesis of a lysosome-targeting Raman probe based on BADY (Lyso-BADY, Fig. 1a) and demonstrate its utility for imaging lysosome in living cells.

|
Download:
|
| Fig. 1. (a) Structures of EdU and Lyso-BADY; (b) SRS intensity of EdU and Lyso-BADY. | |
In this research, morpholine was employed as a lysosome-targeting moiety. Once it entered lysosome, the morpholine group will be protonated and Lyso-BADY can subsequently accumulate in the acidic lysosome matrix. In this way, Lyso-BADY was endowed high hydrophilicity and lysosomal retention. Lyso-BADY was easily synthesized from commercially available methyl 4-formylbenzoate and phenylacetylene (Scheme S1 in Supporting information). The bisarylbutadiynes moiety was constructed by Sonogashira cross-coupling reaction. After the hydrolysis of methyl ester, the benzylic acid was activated by N, N'-disuccinimidyl carbonate and subsequently converted to Lyso-BADY via the addition of 4-(2-aminoethyl) morpholine under mild reaction condition.
In the Raman spectra (Fig. 1b), the signal peak of Lyso-BADY is at 2225 cm-1 and shows a relative Raman intensity versus EdU (RIE) as high as 28 due to phenyl ring enhancement, which is in line with the previous report [27, 28]. It is worth mentioning that the bandwidth (full width at half maximum, FWHM) of Lyso-BADY was about 18 cm-1, and the narrower FWHM means less space will be occupied in the spectrum, which indicates more molecules with different Raman shift can be differentiated in the spectrum with less overlap. In addition, probes with narrower FWHM facilitate multi-color imaging.
After structure confirmation, the photophysical property of Lyso-BADY was further investigated. As shown in Fig. S1 (Supporting information), Lyso-BADY exhibited three absorption band at 297 nm, 316 nm, 338 nm which were red shifted than phenylacetylene precursor, and the red shift can be attributed to the extension of π-conjugation.
To evaluate whether the Raman probe can serve as a lysosomal probe for living cell imaging, the biocompatibility of the probe is of concern. The cytotoxicity of Lyso-BADY by CCK-8 assay in HeLa cell was firstly tested. The results illustrated that Lyso-BADY is of scarce cytotoxic effect in the concentration range from 5 μmol/L to 100 μmol/L, and HeLa cell keeps its original form even at high concentration of Lyso-BADY (Fig. S2 in Supporting information).
Next, we investigated the potentiality of Lyso-BADY as a living cell imaging probe. HeLa cells were incubated in presence of different concentration of Lyso-BADY. At the same time, the commercial lysosomal probe, Lyso-Tracker Red (LTR), was applied as a colocalization fluorescent dye to verify the specificity of our designed molecule. After incubating for 30 min, cells were then subjected to fluorescence and SRS imaging with no precipitation disturbance during the cellular experiment. As presented in Fig. 2a and Fig. S3 (Supporting information), the distributions of C–H bond in lipid droplets and Lyso-BADY are clearly observed at 2885 cm-1 and 2225 cm-1, respectively. It could be easily spotted that the absence of Lyso-BADY led to no Raman signal in HeLa cells when excited by a laser beam. However, the increase of incubating concentration directly accounted for the rising of Raman signal intensity in HeLa cells (Fig. 2b). Specifically, the relative signal intensity was 5 when the concentration of Lyso-BADY was 10 μmol/L, but it reached 9 at 20 μmol/L and 10 at 50 μmol/L, respectively. On the other hand, the merge picture firmly illustrated that the SRS image of Lyso-BADY overlapped perfectly with the fluorescence image of LTR. In addition, the fluorescence signal intensity profile of LTR was found to be highly correlated with the corresponding Raman signal intensity profile (Fig. S5 in Supporting information). These SRS images firmly proved that Lyso-BADY could selectively accumulate at lysosomes of cytoplasm, which means Lyso-BADY can be applied as a probe for lysosomal imaging in living cell with high specificity.

|
Download:
|
| Fig. 2. Sequential SRS imaging of Lyso-BADY and fluorescence imaging of LTR in live cells. (a) HeLa cells were co-incubated with different concentrations of Lyso-BADY (30 min) and Lyso-Tracker Red (200 nmol/L, 30 min). SRS images showed the distributions of C–H at 2885 cm-1 (blue) and Lyso-BADY at 2225 cm-1 (green) and fluorescence images showed it of LTR in live HeLa cells (red). Area of colocalization in the merged image appeared yellow. (b) The Raman signal intensity profile of circled area in Lyso-BADY images. Scale bar is 20 μm. | |
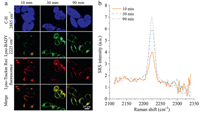
To investigate whether the imaging quality depends on the incubation time, HeLa cells were cultured with 20 μmol/L LysoBADY within various incubation time (10 min, 30 min, 90 min). As shown in Fig. 3, the contrast of Lyso-BADY could be observed within 10 min, indicating rapid ingestion by the cells, and the signal remained a steady state in 30 min, maintaining relatively stable for at least 90 min. The distribution and intensity of LysoBADY signal was further confirmed by intensity profile method (Fig. S4 in Supporting information).
|
Download:
|
| Fig. 3. Sequential SRS imaging of Lyso-BADY and fluorescence imaging of LTR in live cells. (a) HeLa cells were co-incubated with Lyso-BADY (20 μmol/L) at various time and Lyso-Tracker Red (200 nmol/L, 30 min). (b) The SRS spectra of circled area in Lyso-BADY images. Scale bar is 20 μm. | |
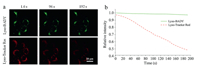
Photostability is considered as one of the key factors in evaluating the performance of cell imaging probes. The photostability of Lyso-BADY was investigated by monitoring the Raman signal intensity via SRS microscopy after being continuously scanned by laser. Meanwhile, LysoTracker assay was carried out utilizing the fluorescence imaging as a contrast. In our test, after 180 s scanning, fluorescence intensity was 50% off, whereas Raman signal intensity remained almost invariable during this time (Fig. 4). What is more, one hour's exposure to laser irradiation totally erased the fluorescence signal of LysoTracker but the left signal of Lyso-BADY was steady and strong (Fig. S6 in Supporting information). This may be attributed to the fact that the diyne moiety was photostable and did not decay upon irradiation for a prolonged time. This feature makes it a valuable alternative for continuous cell imaging since photobleaching effect of fluorophore is problematic for fluorescence imaging.
|
Download:
|
| Fig. 4. Photostability assay of Lyso-BADY and LTR for cellular imaging. (a) SRS images (green) of Lyso-BADY and fluorescence images (red) of LTR at different imaging time. (b) Relative intensity of Lyso-BADY and LTR at different imaging time. Scale bar is 20 μm. | |
In summary, we have synthesized and characterized a novel lysosome-targeting Raman probe called Lyso-BADY and applied it to lysosomal imaging of living cell. It boasts of a Raman signal intensity 28 times stronger than that of EdU because the bisphenyl-substituted diyne intensify the vibration of alkyne dramatically. In addition, the signal of Lyso-BADY locates at 2225 cm-1, the Raman-silent region of cells, which means the Raman images are definitely free from signal disturbance of endogenous molecules. Lyso-BADY also possessed features of nontoxicity, good specificity and high photostability. All these merits render molecules of the same structure as potentially versatile probe for imaging and analysis of organelle or other specific cellular and tissue domain when conjugated with other locating moieties. This work will potentially promote the development of novel Raman probes with varied intracellular targeting with development of the SRS microscopy.
AcknowledgmentWe thank the National Natural Science Foundation of China (Nos. 21432008, 91753201 and 21721005) for financial support.
Appendix A. Supplementary dataSupplementary material related to this article can be found, in the online version, at doi: https://doi.org/10.1016/j.cclet.2019.03.046.
| [1] |
L. Wei, Z. Chen, L. Shi, et al., Nature 544 (2017) 465-470. DOI:10.1038/nature22051 |
| [2] |
L. Wei, F. Hu, Y. Shen, et al., Nat. Methods 11 (2014) 410-412. DOI:10.1038/nmeth.2878 |
| [3] |
Z. Zhao, Y. Shen, F. Hu, W. Min, Analyst 142 (2017) 4018-4029. DOI:10.1039/C7AN01001J |
| [4] |
C.W. Freudiger, W. Min, B.G. Saar, et al., Science 322 (2008) 1857-1861. DOI:10.1126/science.1165758 |
| [5] |
H. Lei, Y. Hu, G. Li, Chin. Chem. Lett. 29 (2018) 509-512. DOI:10.1016/j.cclet.2017.08.012 |
| [6] |
H. Zhang, L. Sun, Y. Zhang, et al., Chin. Chem. Lett. 29 (2018) 981-984. DOI:10.1016/j.cclet.2017.10.017 |
| [7] |
F. Hu, C. Zeng, R. Long, et al., Nat. Methods 15 (2018) 194-200. DOI:10.1038/nmeth.4578 |
| [8] |
L. Wei, F. Hu, Z. Chen, Acc. Chem. Res. 49 (2016) 1494-1502. DOI:10.1021/acs.accounts.6b00210 |
| [9] |
Z. Chen, D.W. Paley, L. Wei, et al., J. Am. Chem. Soc. 136 (2014) 8027-8033. DOI:10.1021/ja502706q |
| [10] |
Z.L. Song, Z. Chen, X. Bian, et al., J. Am. Chem. Soc. 136 (2014) 13558-13561. DOI:10.1021/ja507368z |
| [11] |
L. Wei, Y. Shen, F. Xu, et al., ACS Chem. Biol. 10 (2015) 901-908. DOI:10.1021/cb500787b |
| [12] |
H. Yamakoshi, K. Dodo, M. Okada, et al., J. Am. Chem. Soc. 133 (2011) 6102-6105. DOI:10.1021/ja108404p |
| [13] |
F. Hu, Z. Chen, L. Zhang, et al., Angew. Chem. Int. Ed. 54 (2015) 9821-9825. DOI:10.1002/anie.201502543 |
| [14] |
L. Wei, Y. Yu, Y. Shen, M.C. Wang, W. Min, Proc. Natl. Acad. Sci. U. S. A. 110 (2013) 11226-11231. DOI:10.1073/pnas.1303768110 |
| [15] |
L. Lin, X. Tian, S. Hong, Angew. Chem. Int. Ed. 52 (2013) 7266-7271. DOI:10.1002/anie.201301387 |
| [16] |
Y. Shen, Z. Zhao, L. Zhang, Proc. Natl. Acad. Sci. U. S. A. 114 (2017) 13394-13399. DOI:10.1073/pnas.1712555114 |
| [17] |
A.C. Johansson, H. Appelqvist, C. Nilsson, et al., Apoptosis 15 (2010) 527-540. DOI:10.1007/s10495-009-0452-5 |
| [18] |
C. Settembre, A. Fraldi, D.L. Medina, A. Ballabio, Nature Rev. Mol. Cell Biol. 14 (2013) 283-296. DOI:10.1038/nrm3565 |
| [19] |
H. Xu, D. Ren, Annu. Rev. Physiol. 77 (2015) 57-80. DOI:10.1146/annurev-physiol-021014-071649 |
| [20] |
F.M. Platt, B. Boland, A.C. van der Spoel, J. Cell Biol. 199 (2012) 723-734. DOI:10.1083/jcb.201208152 |
| [21] |
B.L. Wei, P.W. Denton, E. O'Neill, et al., J. Virol. 79 (2005) 5705-5712. DOI:10.1128/JVI.79.9.5705-5712.2005 |
| [22] |
X.J. Cao, L.N. Chen, X. Zhang, et al., Anal. Chim. Acta 920 (2016) 86-93. DOI:10.1016/j.aca.2016.03.029 |
| [23] |
M. Li, H. Ge, V. Mirabello, et al., Chem. Commun. (Camb.) 53 (2017) 11161-11164. DOI:10.1039/C7CC05166B |
| [24] |
X.L. Shi, G.J. Mao, X.B. Zhang, et al., Talanta 130 (2014) 356-362. DOI:10.1016/j.talanta.2014.07.030 |
| [25] |
X. Zhou, F. Su, H. Lu, et al., Biomaterials 33 (2012) 171-180. DOI:10.1016/j.biomaterials.2011.09.053 |
| [26] |
J.A. Mindell, Annu. Rev. Physiol. 74 (2012) 69-86. DOI:10.1146/annurev-physiol-012110-142317 |
| [27] |
H. Yamakoshi, K. Dodo, A. Palonpon, et al., J. Am. Chem. Soc. 134 (2012) 20681-20689. DOI:10.1021/ja308529n |
| [28] |
J. Zhang, S. Yan, Z. He, et al., J. Phys. Chem. Lett. 9 (2018) 4679-4685. DOI:10.1021/acs.jpclett.8b01991 |
 2019, Vol. 30
2019, Vol. 30 

