b Laboratory for Marine Drugs and Bioproducts of Pilot National Laboratory for Marine Science and Technology, Qingdao 266003, China
Marine-derived microorganisms have become an important source of new natural products with various biological activities [1]. In the past decade, more than 4000 new compounds have been isolated from this resource. These account for approximately onethird of the total marine natural products [2]. The microbes associated with marine animals and plants have attracted a lot of attention due to the diversity of species and the novelty of metabolites [3]. Many new compounds with novel structures and significant activities have been isolated from the marine endophytic microorganisms represented by mangrove-associated fungi [4] and algae-associated fungi [5].
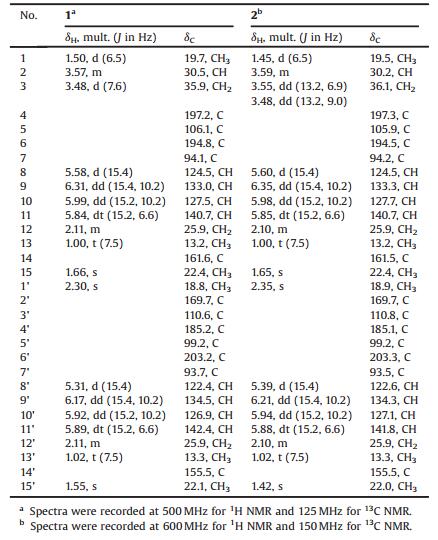
Our laboratory has been working on the bioactive secondary metabolites of marine-derived microorganisms for 15 years [6]. Strains isolated from marine algae, such as Enteromorpha sp., are important objects of our research [7]. So far 40 bioactive natural products have been reported from the Enteromorpha-derived microorganisms. Examples included the anti-inflammatory pleosporallins A–G [8] and conioimide [9], tyrosinase inhibitory myrothenone A [10], antimicrobial pleosporallins D and E [8] and conioscleroderolide [11], as well as cytotoxic penochalasins A–H [12, 13], penostatins A–I [14-16], and communesins A and B [17]. Previously, we have identified two novel methylene-bridged dimers, wailupemycins H and I as well as four new polyketides, 3-O-methylwailupemycin G, and wailupemycins J–L from an actinobacterial strain, Streptomyces sp. OUCMDZ-3434 associated with E. prolifera [18, 19]. Wailupemycins H and I showed significant inhibitory activity against α-glucosidase [18]. In addition, a new cytotoxic pyrrolidinoindoline diketopiperazine dimer, cristatumin E, was isolated from a fungus derived from E. prolifera [20]. As part of our ongoing research of new bioactive compounds from the marine algae-derived microorganisms, 61 fungal strains isolated from E. prolifera, were screened. And the extract of the culture of Paraconiothyrium sp. OUCMDZ-3316 exhibited antibacterial activity. Chemical investigation led to the isolation of two new epimeric polyketide dimers, pafuranones A (1) and B (2) (Fig. 1), together with two known monomers, 3-furanone derivatives, nivefuranone A (3) and 7, 8-dihydronivefuranone A (4) (Table S2 in Supporting information) [21]. The novel dimeric skeleton of compounds 1 and 2 might be formed from the conjugate addition of two monomeric units.
|
Download:
|
| Fig. 1. The structures of compounds 1–4. | |
Pafuranone A (1) was obtained as a colorless oil. Its molecular formula was determined as C30H34O8 based on the HRESIMS peak at m/z 523.2316 [M+H]+ (Fig. S1 in Supporting information), indicating 14 degrees of unsaturation. The 13C NMR spectrum (Fig. S3 in Supporting information) as twelve non-protonated carbons, eight olefinic and one non-oxygenated sp3 methine carbons, three methylene carbons, and six methyl carbons (Table 1). The 1H NMR data (Table 1, Fig. S2 in Supporting information) gave six methyl groups at δH 1.55 (s), 1.66 (s), 2.30 (s), 1.50 (d, J = 6.5 Hz), 1.02 (t, J = 7.5 Hz) and 1.00 (t, J = 7.5 Hz). In addition, the 1H NMR spectrum showed eight olefinic protons at δH 5.31 (d, J = 15.4 Hz), 5.58 (d, J = 15.4 Hz), 5.84 (dt, J = 15.4, 6.6 Hz), 5.89 (dt, J = 15.4, 6.6 Hz), 5.92 (dd, J = 15.4, 10.2 Hz), 5.99 (dd, J = 15.4, 10.2 Hz), 6.17 (dd, J = 15.4, 10.2 Hz) and 6.31 (dd, J = 15.4, 10.2 Hz) (Table 1), which could be attributed to two conjugated diene moieties combined with the COSY spectrum (Fig. 2, Figs. S6 and S7 in Supporting information). The large vicinal coupling constants suggested that all the double bonds were E- configurations.
|
|
Table 1 1H and 13C NMR data for compounds 1 and 2 in CDCl3. |
|
Download:
|
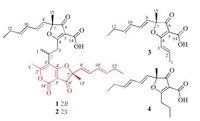
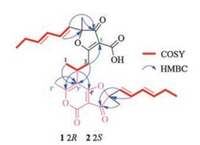
| Fig. 2. Key 2D NMR correlations for the structural assignment of compounds 1 and 2. | |
Comparison of NMR data with those of the known nivefuranone A (3) [21] revealed that pafuranone A (1) contains a similar furanone core and a (1E, 3E)-hexa-1, 3-diene moiety, which could be confirmed by the COSY correlations of H-8/H-9/H-10/H-11/H2-12/H3-13 (Figs. S6 and S7) and the key HMBC correlations of H-9 to C-7, H-8 to C-6/C-15, and H-15 to C-6 (Fig. 2, Figs. S8–S13 in Supporting information). Furthermore, another similar moiety containing a 4H-furo[3, 2-c]pyran-3, 4(2H)-dione dicyclo skeleton could be deduced by the COSY and HMBC correlations (Fig. 2). All these data let us speculate that compound 1 may be a dimer derived from nivefuranone A (3) or its analogue. Two monomeric units were connected through a C-2–C-3 sigma bond, which could be confirmed by the COSY correlations of H3-1/H-2/H2-3 and the HMBC correlations of H-3 to C-5/C-3', H-2 to C-2'/C-4' and H-1/H3-1' to C-3' (Fig. 2, Figs. S6–S13). Considering the 14 degrees of unsaturation and one remaining non-protonated carbon (δC 155.5), a 2-pyrone unit was further confirmed.
The molecular formula of pafuranone B (2) was also determined as C30H34O8 based on the HRESIMS peak at m/z 523.2313 [M+H]+ (Fig. S14 in Supporting information), suggesting that it is an isomer of compound 1. Careful analysis of its NMR data (Table 1, Figs. S15–S18 in Supporting information) and comparison of them with those of compound 1 suggested some minor differences of NMR data for H3-1, H2-3, and H3-1'. 2D NMR data (Fig. 2, Figs. S19–S25 in Supporting information) indicated that compounds 1 and 2 have the same planar structure. We deduced that compound 2 was a 2-epimer of compound 1 through the differences of NMR data together with the analysis of possible biosynthetic pathway.
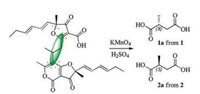
To determine the absolute configurations of C-2, the oxidative cleavage reactions for compounds 1 and 2 were carried out using the acidic potassium permanganate (Fig. 3). (R)-Methylsuccinic acid (1a) and (S)-methylsuccinic acid (2a) were respectively obtained from compounds 1 and 2. The structures of 1a and 2a were confirmed by MS and 1H NMR (Fig. S26–S29 in Supporting information). The specific rotation of 1a ([a]D25 +12.6, H2O) and 2a ([a]D25 -13.3, H2O) match those of (R)- and (S)-methylsuccinic acid in the literatures ([a]D23 +10, H2O) [22] and ([a]D25 -8.7, H2O) [23], respectively. Thus, the absolute configurations of C-2 for compounds 1 and 2 could be determined as R- and S-, respectively.
|
Download:
|
| Fig. 3. Oxidative cleavage of compounds 1 and 2 to methylsuccinic acid. | |
In order to further figure out the configurations of C-7 and C-7', a plausible biosynthetic pathway for pafuranones A (1) and B (2) was postulated (Scheme 1). Both of them might be derived from the same precursor, nivefuranone A (3). Compound 3 undergoes hydrolysis and oxidation to form the intermediate 3a, which further undergoes 1, 6-addition with one molecule of 3 to form the intermediate 3b. By a keto-enol tautomerism, intermediate 3b was then transformed to the intermediate 3c that undergoes a lactonization to generate the 2-pyrone ring. Because this 1, 6-addition lacks stereoselectivity, two epimers (1 and 2) were produced. And all the reactions do not deal with the chiral centre of the monomer (3). Thus, the absolute configurations at C-7 and C-7' of compounds 1 and 2 were deduced the same as compound 3 from the biosynthetic pathway.

|
Download:
|
| Scheme 1. Speculated biosynthetic pathway of 1 and 2. | |
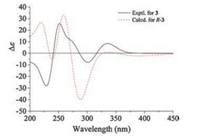
Therefore, we used octant rule [24] for cyclopentenone to determine the C-7 absolute configuration of 3 (Fig. 4). The conformation of (R)-3 as shown in Fig. 4 was placed in the accordance with the octant rule model. The functional group of hexa-1, 3-diene lying in the back upper left area was responsible for the positive Cotton effect which was consistent with the Cotton effect at 335 nm (Δε +8.8). Furthermore, the calculated ECD spectrum of (R)-3 was obtained by the TDDFT [B3LYP/6-31 G(d)] method [25], which was in agreement with the measured ECD curve (Fig. 5), indicating the absolute configuration of compound 3 as R-.

|
Download:
|
| Fig. 4. The octant rule for determining the absolute configuration of compound 3. | |
|
Download:
|
| Fig. 5. Experimental and calculated ECD spectra of compound 3. | |
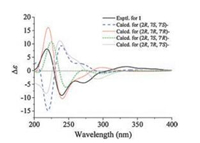
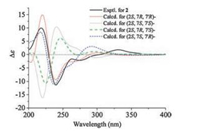
All these results suggested that the absolute configurations of compounds 1 and 2 might be (2R, 7R, 7'R)- and (2S, 7R, 7'R)-, respectively. To confirm the conclusion, the calculated ECD spectra for (2R, 7R, 7'R)-, (2R, 7R, 7'S)-, (2S, 7R, 7'R)-, (2S, 7R, 7'S)-, and their corresponding enantiomers (Table S1) were obtained by the TDDFT [B3LYP/6-31 G(d)] method [25]. The results showed that the experimental ECD spectra of compounds 1 (Fig. 6) and 2 (Fig. 7) matched with the calculated ECD curve of (2R, 7R, 7'R)- and (2S, 7R, 7'R)-, respectively. Thus, the absolute configurations of pafuranones A (1) and B (2) were fully determined.
|
Download:
|
| Fig. 6. Experimental and calculated ECD spectra of compound 1. | |
|
Download:
|
| Fig. 7. Experimental and calculated ECD spectra of compound 2. | |
Compounds 1–4 were evaluated for their antimicrobial activities against Staphylococcus aureus ATCC6538, Pseudomonas aeruginosa ATCC10145, and Candida albicans ATCC10231. However, they did not exhibit antimicrobial activity at the concentration of 0.1 mg/mL (Inhibition zones diameter of positive drug: Ciprofloxacin, 2.1 cm, 1.2 cm; Ketoconazole, 1.5 cm).
The rare genus Paraconiothyrium has been described mainly as endophytes [26] or phytopathogens [27] associated with plants. Other lifestyles like mycoparasitism [28, 29], saprophytism in soil [30] and estuaries [28, 31], association with the digestive tract of insects [32, 33], and opportunistic pathogenesis in humans [34] have also been reported. Studies on the Paraconiothyrium sp. have shown their ample biological potential, including the production of antibiotics [35, 36], anti-inflammatory agents [37], cytotoxic agents [32, 33, 38-41], growth promoting agents [42] and extracellular laccase [43]. These substances were characterized as epoxyphomalins [31], terphenyls [33], phenols [37], and terpenoids [39-41, 44, 45]. This is the first time to study the natural products of Paraconiothyrium sp. from the marine niche. The two novel incomplete symmetry dimeric of furanone and pyrofuranone bear a highly flexible skeleton and rare connection site, which may provide new ideas for metabolic mechanism study and the broad pharmacological screening for green tide prevention, farming and animal husbandry or even human health.
In summary, we identified two new epimeric polyketide dimers from the Enteromorpha-associated fungal strain, Paraconiothyrium sp. OUCMDZ-3316. Only three dimers possessing a similar skeleton, miniolins A–C, have been reported [46]. Although no antimicrobial activity was observed, the novel dimeric structures will provide an insight into the dimerization type of furanone derivatives.
AcknowledgmentsThis work was financially supported by the National Natural Science Foundation of China (Nos. 41876172, 41806086, U1501221, 41376148 & U1606403), and the Fundamental Research Funds for the Central Universities (No. 201841006).
Appendix A. Supplementary dataSupplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.cclet.2019.01.034.
| [1] |
(a) A. Debbab, A.H. Aly, W.H. Lin, P. Proksch, Microb. Biotechnol. 3(2010) 544-563; (b) H. Zhang, Z. Zhao, H. Wang, Mar. Drugs 15(2017) 68; (c) E. Tortorella, P. Tedesco, F.P. Esposito, et al., Mar. Drugs 16(2018) 355. |
| [2] |
(a) J.W. Blunt, A.R. Carroll, B.R. Copp, et al., Nat. Prod. Rep. 35(2018) 8-53; (b) J.W. Blunt, B.R. Copp, R.A. Keyzers, et al., Nat. Prod. Rep. 34(2017) 235-294; (c) J.W. Blunt, B.R. Copp, R.A. Keyzers, et al., Nat. Prod. Rep. 33(2016) 382-431; (d) J.W. Blunt, B.R. Copp, R.A. Keyzers, et al., Nat. Prod. Rep. 32(2015) 116-211; (e) J.W. Blunt, B.R. Copp, R.A. Keyzers, et al., Nat. Prod. Rep. 31(2014) 160-258; (f) J.W. Blunt, B.R. Copp, R.A. Keyzers, et al., Nat. Prod. Rep. 30(2013) 237-323; (g) J.W. Blunt, B.R. Copp, R.A. Keyzers, et al., Nat. Prod. Rep. 29(2012) 144-222; (h) J.W. Blunt, B.R. Copp, M.H.G. Munro, et al., Nat. Prod. Rep. 28(2011) 196-268; (i) J.W. Blunt, B.R. Copp, M.H.G. Munro, et al., Nat. Prod. Rep. 27(2010) 165-237; (j) J.W. Blunt, B.R. Copp, W.P. Hu, et al., Nat. Prod. Rep. 26(2009) 170-244. |
| [3] |
(a) J. Kjer, A. Debbab, A.H. Aly, P. Proksch, Nat. Protoc. 5(2010) 479-490; (b) H. Gao, G. Li, H.-X. Lou, Molecules 23(2018) 646. |
| [4] |
S.K. Deshmukh, M.K. Gupta, V. Prakash, M.S. Reddy, J. Fungi 4 (2018) 101. DOI:10.3390/jof4030101 |
| [5] |
P. Zhang, X. Li, B.G. Wang, Planta Med. 82 (2016) 832-842. DOI:10.1055/s-00000058 |
| [6] |
(a) P. Fu, F. Kong, X. Li, Y. Wang, W. Zhu, Org. Lett. 16(2014) 3708-3711; (b) C. Wang, L. Guo, J. Hao, L. Wang, W. Zhu, J. Nat. Prod. 79(2016) 2977-2981; (c) T. Zhu, Z. Lu, J. Fan, et al., J. Nat. Prod. 81(2018) 2-9; (d) Y. Fan, Y. Wang, P. Fu, et al., Org. Chem. Front. 5(2018) 2835-2839. |
| [7] |
(a) K. Sun, G. Zhu, J. Hao, Y. Wang, W. Zhu, Tetrahedron 74(2018) 83-87; (b) Y. Du, J. Sun, Q. Gong, et al., J. Agric. Food Chem. 66(2018) 1807-1812. |
| [8] |
C.J. Chen, Y.Q. Zhou, X.X. Liu, et al., Tetrahedron Lett. 56 (2015) 6183-6189. DOI:10.1016/j.tetlet.2015.09.079 |
| [9] |
M.F. Elsebai, M. Nazir, S. Kehraus, et al., Eur. J. Org. Chem. 2012 (2012) 6197-6203. DOI:10.1002/ejoc.201200700 |
| [10] |
X. Li, M.K. Kim, U. Lee, et al., Chem. Pharm. Bull. 53 (2005) 453-455. DOI:10.1248/cpb.53.453 |
| [11] |
M.F. Elsebai, S. Kehraus, U. Lindequist, et al., Org. Biomol. Chem. 9 (2011) 802-808. DOI:10.1039/C0OB00625D |
| [12] |
C. Iwamoto, T. Yamada, Y. Ito, K. Minoura, A. Numata, Tetrahedron 57 (2001) 2997-3004. DOI:10.1016/S0040-4020(01)00153-3 |
| [13] |
A. Numata, C. Takahashi, Y. Ito, et al., J. Chem. Soc. Perkin Trans. 1 (1996) 239-245. |
| [14] |
C. Iwamoto, K. Minoura, T. Oka, et al., Tetrahedron 55 (1999) 14353-14368. DOI:10.1016/S0040-4020(99)00884-4 |
| [15] |
C. Iwamoto, K. Minoura, S. Hagishita, K. Nomoto, A. Numata, Tetrahedron 3 (1998) 449-456. |
| [16] |
C. Takahashi, A. Numata, T. Yamada, et al., Tetrahedron Lett. 37 (1996) 655-658. DOI:10.1016/0040-4039(95)02225-2 |
| [17] |
A. Numata, C. Takahashi, Y. Ito, et al., Tetrahedron Lett. 34 (1993) 2355-2358. DOI:10.1016/S0040-4039(00)77612-X |
| [18] |
Z. Chen, J. Hao, L. Wang, et al., Sci. Rep. 6 (2016) 20004. DOI:10.1038/srep20004 |
| [19] |
H. Liu, Z. Chen, G. Zhu, et al., Tetrahedron 73 (2017) 5451-5455. DOI:10.1016/j.tet.2017.07.052 |
| [20] |
Y. Li, K.L. Sun, Y. Wang, et al., Chin. Chem. Lett. 24 (2013) 1049-1052. DOI:10.1016/j.cclet.2013.07.028 |
| [21] |
(a) Y. Shiono, T. Hatakeyama, T. Murayama, T. Koseki, Nat. Prod. Commun. 7(2012) 1065-1068; (b) M. Vallet, Q.P. Vanbellingen, T. Fu, et al., J. Nat. Prod. 80(2017) 2863-2873 (c) T. Fujiwara, A. Sato, Y. Kawamura, K. Matsumoto, H. Itazaki, Patent, JP 6239852, 1994. |
| [22] |
V.H.T. James, J. Chem. Soc. (1955) 637-639. DOI:10.1039/jr9550000637 |
| [23] |
K. Weinges, E. Paulus, Eur. J. Org. Chem. 681 (1965) 154-161. |
| [24] |
(a) von C. Djerassi, Optical Rotatory Dispersion: Applications to Organic Chemistry, McGraw-Hill, New York, 1960; (b) L. Velluz, M.L. Grand, M. Grosjean, Optical Circular, Dichroism: Principles, Measurements, and Applications, Academic Press, New York, 1965; (c) S. Lin, T. Shi, K. Chen, et al., Chem. Commun. 47(2011) 10413-10415; (d) P. Fu, J.B. MacMillan, Org. Lett. 17(2015) 3046-3049; (e) P. Fu, S. La, J.B. MacMillan, J. Nat. Prod. 79(2016) 455-462. |
| [25] |
P.J. Stephens, J.J. Pan, K. Krohn, J. Org. Chem. 72 (2007) 7641-7649. DOI:10.1021/jo071183b |
| [26] |
Y.K. Zheng, X.G. Qiao, C.P. Miao, et al., Ann. Microbiol. 66 (2016) 529-542. DOI:10.1007/s13213-015-1153-7 |
| [27] |
M. Cloete, P.H. Fourie, U. Damm, P.W. Crous, L. Mostert, Phytopathol. Mediterr. 50 (2011) 176-190. |
| [28] |
G.J.M. Verkley, M. da Silva, D.T. Wicklow, P.W. Crous, Stud. Mycol. 50 (2004) 323-335. |
| [29] |
J.M. Whipps, M. Gerlagh, Mycol. Res. 96 (1992) 897-907. DOI:10.1016/S0953-7562(09)80588-1 |
| [30] |
U. Damm, G.J.M. Verkley, P.W. Crous, et al., Persoonia 20 (2008) 9-17. DOI:10.3767/003158508X286842 |
| [31] |
I.E. Mohamed, S. Kehraus, A. Krick, et al., J. Nat. Prod. 73 (2010) 2053-2056. DOI:10.1021/np100310k |
| [32] |
C.X. Liu, L. Wang, J.F. Chen, et al., Magn. Reson. Chem. 53 (2015) 317-322. DOI:10.1002/mrc.v53.4 |
| [33] |
F. Ren, S. Chen, Y. Zhang, et al., J. Nat. Prod. 81 (2018) 1752-1759. DOI:10.1021/acs.jnatprod.8b00106 |
| [34] |
M.A. Colombier, A. Alanio, B. Denis, et al., J. Clin. Microbiol. 53 (2015) 2084-2094. DOI:10.1128/JCM.00295-15 |
| [35] |
T. Suzuki, N.R. Ariefta, T. Koseki, et al., Fitoterapia 132 (2019) 75-81. DOI:10.1016/j.fitote.2018.11.017 |
| [36] |
C. Anisha, P. Sachidanandan, E.K. Radhakrishnan, Curr. Microbiol. 75 (2018) 343-352. DOI:10.1007/s00284-017-1387-7 |
| [37] |
T.H. Quang, D.C. Kim, P. van Kiem, et al., J. Antibiot. 71 (2018) 826-830. DOI:10.1038/s41429-018-0073-8 |
| [38] |
N. Cho, T.T. Ransom, J. Sigmund, et al., J. Nat. Prod. 80 (2017) 2037-2044. DOI:10.1021/acs.jnatprod.7b00170 |
| [39] |
S. Chen, Y. Zhang, C. Zhao, et al., Fitoterapia 99 (2014) 236-242. DOI:10.1016/j.fitote.2014.09.021 |
| [40] |
S. Chen, Y. Zhang, S. Niu, X. Liu, Y. Che, J. Nat. Prod. 77 (2014) 1513-1518. DOI:10.1021/np500302e |
| [41] |
Y. Shiono, M. Kikuchi, T. Koseki, et al., Phytochemistry 72 (2011) 1400-1405. DOI:10.1016/j.phytochem.2011.04.016 |
| [42] |
X. Li, X. He, L. Hou, et al., Sci. Rep. 8 (2018) 7896. DOI:10.1038/s41598-018-26183-0 |
| [43] |
H. Forootanfar, M.A. Faramarzi, A.R. Shahverdi, M.T. Yazdi, Bioresour. Technol. 102 (2011) 1808-1814. DOI:10.1016/j.biortech.2010.09.043 |
| [44] |
Z. Guo, F. Ren, Y. Che, G. Liu, L. Liu, Molecules 20 (2015) 14611-14620. |
| [45] |
L. Liu, X. Chen, D. Li, et al., J. Nat. Prod. 78 (2015) 746-753. DOI:10.1021/np5009569 |
| [46] |
H.Y. Tang, Q. Zhang, Y.Q. Gao, et al., RSC Adv. 5 (2015) 2185-2190. DOI:10.1039/C4RA11712C |
 2019, Vol. 30
2019, Vol. 30