b Tsinghua-Peking Center for Life Sciences, Ministry of Education Key Laboratory of Bioorganic Phosphorus Chemistry and Chemical Biology, Department of Chemistry, Tsinghua University, Beijing 100084, China;
c Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing 100081 China;
d School of Biological Science, Beijing Institute of Technology, Beijing 100084, China
The ubiquitination system regulates various vital cellular processes in eukaryotic cells [1, 2]. Deubiquitinases (DUBs) are a superfamily of around 100 enzymes that can reverse ubiquitination by hydrolyzing mono- or poly-ubiquitin chain from distinct substrates proteins [3, 4]. Recently, disorder of specific DUBs has been implicated in many serious diseases including cancer, autoimmune diseases, and neurodegenerative disease, etc. [5]. For example, Ubiquitin (Ub) specific protease-14 (USP14) is one of the best characterized diseaserelated DUBs. It has been identified over-expressing in various cancer cell lines and proposed to function by blocking proteasome function and upregulating Wnt/β-catenin pathway [6]. Therefore, USP14 has emerged as an attractive drug target, and the selective inhibition of USP14 has been proposed for future cancer therapy [7-9].
In order to screen and identify specific inhibitors of DUBs, it requires Ub probes within a reporting group on the C-terminus to measure the activity of DUBs [10-12]. Among all the Ub probes, ubiquitin-7-amido-4-methylcoumarin (Ub–AMC) is one of the most widely used reagent in high throughput screening of DUBs inhibitors [13-18], which has been used successfully to find a lead USP14 inhibitor (IU1, IC50 = 4-5 mmol/L) [19]. However, AMC fluorescence is monitored using an excitation wavelength of 345 nm and an emission wavelength of 445 nm. Thus, the candidate compounds that absorb or emit fluorescence at this range may generate false positive results [20, 21]. Moreover, the reaction conditions of Ub-AMC also need to be optimized for each specific application. Due to the false positive results and unsatisfactory repeatability, further optimization and validation of the lead inhibitors requires more reliable probes.
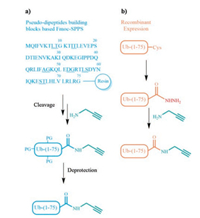
Ub-propargylamide is an activity-based probe that can forms covalent bond with the catalytic cysteine of active DUBs. It has been reported that the measurement of DUBs activity with Ub-PA is more repeatable and credible than that with Ub-AMC.Thus, Ub-PA instead of Ub-AMC is an indispensable tool for USP14 targeting inhibitors validation [22-24]. Previous production of Ub-PA relies on tedious Fmoc-based solid-phase peptide synthesis (SPPS) of Ub(1-75)-OH, after which propargylamine is used to modify the C-terminus of the ubiquitin segment (Scheme 1a) [22, 25]. SPPS and protein ligation methods makes great contributions for the production Ub-based probes.However, the relative low yield of the SPPS route limits the applicationofUb-PA in drug discovery industry.Here, we presented a facile semi-synthesis method for the large quantity production of Ub-PA (Scheme 1b) and demonstrated the utility of our products for USP14 inhibitors validation.
|
Download:
|
| Scheme 1. Strategies for the synthesis of Ub-PA. (a) Pseudo-dipeptides building blocks based solid-phase peptide synthesis route (underlined amino acids represent pseudo-dipeptides); (b) This work: semi-synthesis route. | |
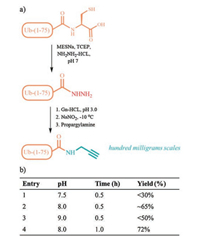
Inspired by recent advances in recombinant ubiquitin-NHNH2 production [26, 27] and subsequent ubiquitin-NHNH2 ligation and modification [28-31], we designed a novel semi-synthetic route to produce Ub-PA, and the protocol takes only two steps of facile chemical reactions. As shown in Fig. 1, recombinant Ub(1-75)-Cys was converted into key intermediate Ub(1-75)-NHNH2, and then converted into Ub-PA by propargylamine aminolysis. In details, the first step is the production of recombinant Ub(1-75)-Cys protein. We performed large-scale E. coli BL21 (DE3) cells protein expression and purification as previous method [16]. Then we treated Ub(1-75)-Cys protein solution with the hydrazinolysis reagent (NH2NH2-HCl 50 mg/mL, Mesna 100 mg/mL, tris(2-chloroethyl) phosphate (TCEP) 5 mg/mL) and react under pH 7 overnight to obtain Ub(1-75)-NHNH2. Crude Ub(1-75)-NHNH2 was then purified by high performance liquid chromatography (HPLC) with around 60% yield and lyophilized into powder in hundred milligrams scale.
|
Download:
|
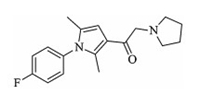
| Fig. 1. Molecular structure of IU1. | |
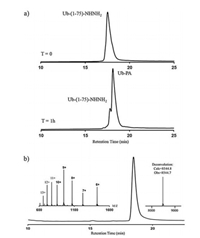
Secondly, Ub(1-75)-NHNH2 powder was dissolved in Guanidinium chloride Buffer (6.0 mol/L Gn-HCL, 0.2 mol/L PBS, pH 3) and oxidized into Ub(1-75)-N3 with NaNO2 under -10 ℃ in ice-salt baths in 20 min. Then, we sought to optimize the reaction conditions for the final propargylamine aminolysis. To this end, propargylamine liquid (5 equiv. to Ub(1-75)-NHNH2) was added into the solution and adjusted to pH 7.5, 8.0, 9.0 separately with hydrochloric acid, then react in room temperature (Fig. 2b). The reaction was monitored by analytical HPLC, and we found that most of Ub(1-75)-NHNH2 could converted into Ub-PA in pH 8.0, and the reaction completed within 1 h (Fig. 3a).
|
Download:
|
| Fig. 2. (a) Semi-synthetic route for Ub-PA: 1. recombinant expression of Ub variants, 2. hydrazinolysis, 3. Propargylamine aminolysis. (b) The optimization of propargylamine aminolysis reactions. | |
|
Download:
|
| Fig. 3. (a) HPLC traces of the aminolysis reaction; (b) HPLC and ESI-MS analysis of purified Ub-PA. | |
After aminolysis of Ub(1-75)-NHNH2, the reaction solution was quenched by the addition of Gn·HCl buffer (6.0 mol/L Gn·HCl, PBS, pH 3.0), then the crude product was purified by semi-preparative HPLC (72% isolated yield). Finally, we characterized the purified Ub-PA by using analytical HPLC and ESI-MS. The chromatogram and spectrogram verified the purity and molecular weight of our semi-synthetic product (Fig. 3b). Overall, the entire production process of the semi-synthetic Ub-PA takes only two days. With cheap recombinant Ub-variants as starting material, we could easily obtain hundred milligrams scales of Ub-PA in high purity. In contrast, traditional SPPS routes is time-consuming and requires a large amount of expensive Pseudo-dipeptides [22, 25]. A whole SPPS batch can only yield less than one milligram Ub-PA, which limit its application in the development of DUBs targeting inhibitors.
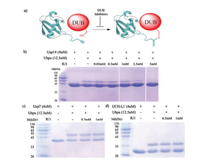
To further test the ability of our semi-synthetic Ub-PA for USP14 inhibitors validation, we firstly recombinantly expressed USP14 and two other DUBs (USP7, UCH-L1) as negative controls. Then we used the probe to evaluate the inhibition of DUBs with IU1 and analyzed the results with sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). In details, GST–tagged USP14 and other DUBs were all expressed in E. coli. strain BL21 (DE3) cells separately. After, cultures were grown at 37 ℃ until OD600 nm reached to 1.0. The protein expression was induced with 0.2 mmol/L IPTG at 22 ℃ for 8-12 h. Subsequently, cells were harvested in lysis buffer (25 mmol/L Tris, 150 mmol/L NaCl) containing PMSF protease inhibitor and lysed by sonification. The cleared lysates were purified by GST Sepharose 4B column (GE healthcare). The GST-tag was removed by PreScission proteases in cleavage buffer (25 mmol/L tris-HCl (pH 8.0), 150 mmol/L NaCl) for 2 h at room temperature. Then, we used AKTA pure for ion exchange chromatography and gel filtration chromatography purification. To test the utility of the Ub-PA probe, we mixed USP14 with its specific inhibitor IU1 in lysis buffer. After incubation for 30 min, added excessive Ub-PA and incubated for 30 min. After SDS-PAGE protein isolation, we found the crosslinking band intensity of USP14 and Ub-PA decreased along the inhibitor concentration gradient (Fig. 4b). Then, we examine the selectivity of the inhibitors with USP7, UCH-L1, and found that the inhibitors show great selectivity towards USP14, while cannot inhibit other DUBs efficiently (Fig. 4c). Together, these results demonstrate the ability of the semi-synthetic Ub-PA as an effective probe for validating USP14 specific inhibitors.
|
Download:
|
| Fig. 4. (a) Diagram of Ub-PA crosslinking with DUB; (b) SDS-PAGE shows that IU1 inhibits USP14 activity in a concentration-dependent manner; (c, d) IU1 cannot inhibit USP7 or UCHL1. | |
In summary, our work enables facile and efficient synthesis of large-scale Ub-PA. The semi-synthetic protocol avoids tedious and specialized protein chemical synthesis techniques, thus can be readily carried out in biological laboratories. The biochemical assays show that our semi-synthetic Ub-PA is an effective probe for the validation of the activity of DUBs inhibitors, and is therefore expected to be further used to optimize and identify potent small molecules targeting USP14 and other diseases-related DUBs. We expect that convenient and repeatable Ub-PA validation assay may help improve the efficiency of DUBs targeting drug discovery.
AcknowledgmentsThis work was supported by the National Natural Science Foundation of China (Nos. 91753205 and 21877024 to Y. Li and No. 31640016 to Z. Mei), the Fundamental Research Funds for the Central Universities (No. PA2017GDQT0021 to Y. Li), the funding of National Key R&D program of China (No. 2017YFD0500400 to Z. Mei), the Fundamental Research Funds for the Central Non-profit Scientific Institution (No. Y2016JC25 to Z. Mei) and the Agricultural Science and Technology Innovation Program to Z.Mei.
Appendix A. Supplementary dataSupplementary material related to this article canbe found, in the online version, at doi:https://doi.org/10.1016/j.cclet.2018.09.003.
| [1] |
D. Komander, M. Rape, Annu. Rev. Biochem. 81 (2012) 203-229. DOI:10.1146/annurev-biochem-060310-170328 |
| [2] |
A. Ciechanover, Nat. Rev. Mol. Cell Biol. 16 (2015) 322-324. DOI:10.1038/nrm3982 |
| [3] |
D. Komander, M.J. Clague, S. Urbé, Nat. Rev. Mol. Cell Biol. 10 (2009) 550-563. DOI:10.1038/nrm2731 |
| [4] |
T.E. Mevissen, D. Komander, Annu. Rev. Biochem. 86 (2017) 159-192. DOI:10.1146/annurev-biochem-061516-044916 |
| [5] |
J.A. Harrigan, X. Jacq, N.M. Martin, et al., Nat. Rev. Drug Discov. 17 (2018) 57-77. |
| [6] |
H. Jung, B.G. Kim, W.H. Han, et al., Oncogenesis 2 (2013) e64. DOI:10.1038/oncsis.2013.28 |
| [7] |
P. D'arcy, S. Brnjic, M.H. Olofsson, et al., Nat. Med. 17 (2011) 1636-1640. DOI:10.1038/nm.2536 |
| [8] |
M.I. Davis, A. Simeonov, Drug Target Rev. 2 (2015) 60-64. |
| [9] |
X. Huang, V.M. Dixit, Cell Res. 26 (2016) 484-498. DOI:10.1038/cr.2016.31 |
| [10] |
Y. Leestemaker, H. Ovaa, Drug Discov. Today Technol. 26 (2017) 25-31. DOI:10.1016/j.ddtec.2017.11.006 |
| [11] |
D.S. Hewings, J.A. Flygare, M. Bogyo, et al., FEBS J. 284 (2017) 1555-1576. DOI:10.1111/febs.14039 |
| [12] |
G.B. Tilburg, A.F. Elhebieshy, H. Ovaa, Opin. Struct. Biol. 38 (2016) 92-101. DOI:10.1016/j.sbi.2016.05.022 |
| [13] |
Y. Liu, H.A. Lashuel, S. Choi, et al., Chem. Biol. 10 (2003) 837-846. DOI:10.1016/j.chembiol.2003.08.010 |
| [14] |
A.P. Turnbull, S. Ioannidis, W.W. Krajewski, Nature 550 (2017) 481-486. DOI:10.1038/nature24451 |
| [15] |
Y.T. Li, Y.C. Huang, Y. Xu, et al., Tetrahedron 72 (2016) 4085-4090. DOI:10.1016/j.tet.2016.05.041 |
| [16] |
G.C. Chu, J.S. Bai, Y.F. Kong, et al., Tetrahedron 74 (2018) 3931-3935. DOI:10.1016/j.tet.2018.05.081 |
| [17] |
C. Reverdy, S. Conrath, R. Lopez, et al., Chem. Biol. 19 (2012) 467-477. DOI:10.1016/j.chembiol.2012.02.007 |
| [18] |
A.H. Mermerian, A. Case, R.L. Stein, et al., Bioorg. Med. Chem. Lett. 17 (2007) 3729-3732. DOI:10.1016/j.bmcl.2007.04.027 |
| [19] |
B.H. Lee, M.J. Lee, S. Park, et al., Nature 467 (2010) 179-181. DOI:10.1038/nature09299 |
| [20] |
C. Ndubaku, V. Tsui, J. Med. Chem. 58 (2014) 1581-1595. |
| [21] |
B. Nicholson, S. Kumar, S. Agarwal, et al., J. Biomol. Screen. 19 (2014) 989-999. DOI:10.1177/1087057114527312 |
| [22] |
R. Ekkebus, S.I. van Kasteren, Y. Kulathu, et al., J. Am. Chem. Soc. 135 (2013) 2867-2870. DOI:10.1021/ja309802n |
| [23] |
A.P. Lawson, D.W. Bak, D.A. Shannon, et al., Oncotarget 8 (2017) 51296. |
| [24] |
T.E. Mevissen, M.K. Hospenthal, P.P. Geurink, et al., Cell 154 (2013) 69-184. |
| [25] |
F. El Oualid, R. Merkx, R. Ekkebus, et al., Angew. Chem. Int. Ed. 49 (2010) 10149-10153. DOI:10.1002/anie.201005995 |
| [26] |
A.L. Adams, B. Cowper, R.E. Morgan, et al., Angew. Chem. Int. Ed. 125 (2013) 13300-13304. DOI:10.1002/ange.201304997 |
| [27] |
L. Xu, J.F. Huang, C.C. Chen, et al., Org. Lett. 20 (2017) 329-332. |
| [28] |
G.M. Fang, Y.M. Li, F. Shen, et al., Angew. Chem. Int. Ed. 50 (2011) 7645-7649. DOI:10.1002/anie.201100996 |
| [29] |
T. Curtius, J. Prakt. Chem. 70 (1904) 57-72. DOI:10.1002/prac.19040700103 |
| [30] |
N. Fujii, H. Yajima, J. Chem. Soc. Perkin Trans. 1 (1981) 831-841. |
| [31] |
J.S. Zheng, S. Tang, Y.K. Qi, et al., Nat. Protoc. 8 (2013) 2483-2495. DOI:10.1038/nprot.2013.152 |
 2019, Vol. 30
2019, Vol. 30 

