b College of Chemistry, Sichuan University, Chengdu 610064, China;
c Beijing National Laboratory for Molecular Science, Center for Molecular Science, State Key Laboratory for Structural Chemistry for Unstable and State Species, Institute of Chemistry, Chinese Academy of Sciences, Beijing 100190, China
Supramolecular assembly of cyanine dyes has aroused widely interest due to their controllable process and wide application [1]. Assembly process of cyanine dyes could be constructed and regulated by biomolecules [2, 3]. Duplex DNAs with proper grooves were widely used as templates to self-assemble cyanine dye into dimers or complex aggregates [4-6]. Besides duplex DNA, DNAs also could form second structures, such as G-quadruplexes and imotifs, which have specific chemical characteristic [7]. These properties might also be as a good template to modulate assembly and disassembly of cyanine dyes.
G-quadruplexes (G4s) are guanine-rich sequences capable of folding into four-stranded nucleic acid structures, composed of planar arrangements of four guanines bases stabilized by eight hoogsteen hydrogen bonds in monovalent cation solution [8, 9]. Grich sequences with high potential to form G4s are found in many important genomic regions, such as telomeres, promoter regions of important oncogenes, gene bodies and untranslated regions [10]. It has been proposed that in vivo the formation of G4s is related to a certain important biological functions. G4s have square planar configuration, grooves and loops. Benefiting from these structural properties, G4s as templates might easily interact with cyanine dyes as duplex DNAs do. Specific G4 structures might regulate assembly and disassembly process of cyanine dyes (Fig. 1).
|
Download:
|
| Fig. 1. The structures of TC and TC-P4. | |
Our previous works had reported that self-assembly and disassembly process of some monomeric cyanine dyes could successfully be regulated by specific G4s [11, 12]. In this strategy, these molecules firstly self-assemble into aggregates, then can be disassembled into monomer by specific G4 template since the binding affinity between these molecules and G4s are stronger than that among molecules in the dye self-assembly accompanied with specific recognition signals. Recently, dimeric ligands through connecting two monomers with a flexible linker used for G4s have been attracted more interest due to good selectivity of G4s [13, 14]. We had designed a series of dimeric cyanine dyes with various linker length and aromatic rings accompanied with good selfassembly and disassembly properties [15, 16]. The dimeric dye (TCP4) with three repeat units (oligo-oxyethylene) linker shows higher recognition ability of specific G4 than these with longer or shorter linker one. In order to clarify the influence of linker on the assembly and disassembly of dimeric cyanine dye, herein, we compared a dimeric cyanine dye (TC-P4) with its corresponding monomer (TC) in self-assembly and disassembly properties with specific DNA templates. TC and TC-P4 could assemble into different aggregates in PBS. These aggregates could be disassembled into dimer and/or monomer using different DNA templates. These different disassembly properties of cyanine dyes treated with specific DNA templates could exhibit specific recognition signals. TC-P4 exhibited more specific absorption spectral changes and great fluorescence enhancement upon binding to parallel c-myc G4 than those of TC. Even in the large amount of duplex DNAs, TC-P4 still presented high binding affinity to G4 in solution.
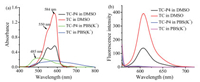
To demonstrate the photophysical properties of TC and TC-P4, the UV–vis spectroscopy and fluorescence spectroscopy of these dyes were performed in solution. It is well known that dye aggregation depends on the self-structure and environment. Some cyanine dyes typically tend to form H-aggregates or J-aggregates in polar solvent, such as aqueous solution [17]. The UV–vis spectra of TC and TC-P4 in DMSO and PBS(K+) (10 mmol/L KH2PO4/K2HPO4, 70 mmol/L KCl, 1 mmol/L EDTA, pH 7.4) are shown in Fig. 2a. In DMSO, TC exhibits a primary absorption band at 584 nm assigned to monomer band, while TC-P4 exhibits a primary absorption at 584 nm assigned to monomer band, as well as a smaller shoulder peak at 550 nm assigned to dimer band, according to reported analogs, such as ETC [18], YOYO-3 [19] and BHmC [20] (Figs. S10 and S11, Table S2 in Supporting information). In PBS(K+), due to the extended planar and positive conjugated system, TC-P4 exhibits a primary absorption band at 480 nm assigned to H-aggregates, while TC forms mixture aggregates. The fluorescence spectra of TC and TC-P4 in DMSO and PBS(K+) were also tested. As shown in Fig. 2b, TC shows a primary absorption band at 607 nm assigned to monomer, while TC-P4 also shows a primary bands at 607 nm assigned to monomer, which is weaker than that of TC. This characteristic might be due to formation of dimer in TC-P4 to cause strong excitonic coupling [17]. TC and TC-P4 exhibit a weak fluorescence emission in PBS(K+). H-aggregates show extensive π-packing that cause a hypsochromic shift of the absorption band and a reduced fluorescence emission due to strong excitonic coupling [17]. The great difference of fluorescence intensity for TC and TC-P4 in solvent could be easily distinguished when aggregates were transformed into dimer and/or monomer.
|
Download:
|
| Fig. 2. (a) UV–vis spectra and (b) fluorescence spectra of 5 mmol/L TC or TC-P4 in DMSO and PBS(K+) (10 mmol/L K2HPO4/ KH2PO4, 70 mmol/L KCl, 1 mmol/L EDTA, pH 7.4). | |
To demonstrate the disassembly process of TC and TC-P4 aggregates, different DNA templates were used to assay. Due to TC and TC-P4 with planar aromatic rings, G-quadruplex (G4) with planar G-tetrads was chosen and might easily interact with planar aromatic rings of TC/TC-P4 to change states of aggregates. The stable conformation of parallel G4 (c-myc) was chosen to induce aggregates transformation. c-myc is an important proto-oncogene involved in cellular proliferation and cell growth. Over-expression of c-myc is also related to various cancers, including breast, colon, cervix, small-cell lung carcinomas, osteosarcomas and myeloid leukemias [21]. Duplex DNAs [D24 (human telomeric), duplex CT (calf thymus, a type of non-telomeric double strand DNA), intramolecular hairpin duplex D26], and single-stranded DNA [S17 (non-telomeric)] were as control groups to interact with TC/ TC-P4. The c-myc could induce TC aggregation into a primary monomer peak at 584 nm; D24, CT, D26 and S17 also could induce the appearance of monomer peak (Figs. 3a and b). This result suggested that TC could bind to all these oligonucleotides in the form of monomer. However, the absorption spectral change trend of TC-P4 with these DNA motifs was not the same as those of TC. TC-P4 treated with c-myc results in the transformation from Haggregates to dimer and monomer; while D24, D26, CT and S17 could not induce the appearance of monomer and/or dimer (Figs. 3c and d). The significant signature (H-aggregates and monomer are separated by about 100 nm) contributed TC-P4 to be a better probe for c-myc G-quadruplex from other DNA motifs.

|
Download:
|
| Fig. 3. (a) Absorption spectra of 5 mmol/L TC in the various concentration of c-myc. (b) Absorption spectra of 5 mmol/L TC in 40 mmol/L c-myc, D26, D24, CT (800 mg/mL) and S17, respectively. (c) Absorption spectra of 5 mmol/L TC-P4 in the various concentration of c-myc. (d) Absorption spectra of 5 mmol/L TC-P4 in 40 mmol/L c-myc, D26, D24, CT (800 mg/mL) and S17, respectively. | |
In order to provide more detail of recognition signal, the fluorescent properties of TC-P4 with various DNA motifs were also carried out at the same time (Figs. S1 and S2 in Supporting information). As shown in Fig. 4a, the spectral change showed two stages. At low [c-myc]/[TC-P4] (less than 1:1) ratio, TC-P4 shows two peaks and partially overlapped fluorescent emission bands at ~587 nm and 602 nm, assigned to dimer and monomer, respectively. TC-P4 dimer showed moderate fluorescence intensity in the presence of c-myc since dimer existed in a rigid and confined environment [22]. At high [c-myc]/[TC-P4] (more than 1:1) ratio, fluorescence was strongly increased by c-myc, which could be reasonably attributed to the restriction of radiation deactivation of the excited singlet state for monomer. Based on these spectral results, it could be considered that the interaction between TC-P4 and c-myc showed two concentration-related stages: TC-P4 Haggregates disassemble into dimer and then dimer disassembles into monomer. In the case of duplex and single-stranded DNAs, as shown in Fig. 4c, the fluorescence intensity of TC-P4 was weakly enhanced, indicated that the weak interaction occurred to these DNA motifs with TC-P4. In contrast, TC bound to c-myc resulted in strong fluorescent enhancement. The fluorescent titration between TC and duplex and single-stranded DNAs were also carried out (Fig. 4d). The fluorescence intensity enhancement of TC treated with duplex and single-stranded DNAs was weaker than that with c-myc. The fluorescence intensity of dyes treated with DNA motif might mainly come from their monomer formation. The enhancement of fluorescence intensity of TC treated with c-myc was larger than that of TC-P4. Duplex and single-stranded DNAs could also induce TC monomer appearance, resulting in a certain fluorescent enhancement; while these DNA motifs could not induce the appearance of TC-P4 monomer, accompanying with relatively weak fluorescent enhancement. The relative fluorescent selectivity of TC-P4 for G-quadruplex via duplex or single-stranded DNAs was larger than that of TC. Thus, TC-P4 exhibited higher relative fluorescent selectivity for c-myc over duplex and single-stranded DNAs than that of TC (Figs. 4c and d).

|
Download:
|
| Fig. 4. The fluorescence intensity of TC-P4 (a) and TC (b) in the presence of various concentration of c-myc. Fluorometric titration curves of TC-P4 at 609.6 nm (c) or TC at 605.0 nm (d) with different DNA structures, including G-quadruplex (c-myc), duplex DNAs (D24, D26 and CT) and single-stranded DNA (S17). | |
It is well known that in vivo, except for the single-stranded Grich telomeric 30-overhang, most of the putative G-quadruplexforming sequences are embedded in double-stranded genome [23]. CT is a type of non-telomeric double strand DNA, which is considered as a mixture of various duplex DNAs with relatively random sequences. To evaluate the specificity of TC-P4 to G4, the competition binding assay was studied (Fig. S3 in Supporting information). TC-P4 or TC (1.5 mmol/L) was added into the mixture of 3 mmol/L c-myc and different concentrations of CT. In the presence of 250 mg/mL CT, the fluorescence spectra of TC-P4 with c-myc was nearly the same as that without CT, while at 200 mg/mL of TC was nearly the same as that without CT. TC-P4 also presents higher binding affinity to c-myc in the large excess of duplex D26 than that of TC (Fig. S3). The results indicated that duplex DNAs (CT or D26), even in a large amount, could not influence on the disassembly of TC-P4 aggregates with c-myc.
The colorimetric test in solution was carried out (Fig. S4 in Supporting information). 10 mmol/L TC/TC-P4 was dissolved in PBS (K+), and then different DNA templates were added, respectively. TC in PBS(K+) was nearly colorless. TC in PBS(K+) treated with cmyc could make color change and show the pink color, while other duplex and single-stranded DNAs could not cause the same color change. The color of TC-P4 in PBS(K+) treated with c-myc also showed difference from TC-P4 treated with other DNA templates. TC showed better color discrimination ability of c-myc from duplex and single-stranded DNAs than that of TC-P4 by naked eyes. This result could be come from two reasons: 1) H-aggregates of TC-P4 in PBS(K+) shows darker color than that of TC; 2) Fluorescence intensity of TC treated with 40 mmol/L c-myc is stronger than that of TC-P4.
Organic ligands bind to G4s mainly through various binding mode, such as end-stacking and groove binding [24, 25]. The endstaking mode on parallel G4s are usually more feasible. Our previous work had proved that TC-P4 could interact with c-myc in the form of monomer like a clip-like mode [16]. Here, in order to confirm TC binding mode, a competition inhibition experiment on the G4/hemin peroxidase activity by TC was investigated (Fig. S6 in Supporting information) [26]. Hemin binding to G4 by endstacking mode was considered to possess catalytic activity on the oxidation of ABTS2- by H2O2. The cyanine dye with the same binding sites will compete with hemin for G4 binding, which result in decreasing the rate of catalytic oxidation of G4/hemin peroxidase. The parallel G4 (c-myc) was used to evaluate the inhibition activity of TC on G4/hemin peroxidase. As we expected, c-myc exhibited strong peroxidase activities in the absence of TC, and peroxidase activity was strongly inhibited in the presence of TC, indicating that TC strongly competed with hemin to bind to the end of G-quartet.
To clarify cyanine dye for the discrimination ability of different types of G4s, fluorescent and absorption titration of TC-P4 with other types of G4 motifs were also carried out. These DNA motifs include intramolecular parallel-stranded G4s (myc22 and c-kit1), intermolecular parallel G4 (H7), hybrid G4s (M24, bcl-2 2345 and H24) and antiparallel-stranded G4s (A22, A24 and TBA) [15]; RNA G4s including parallel-stranded G4s (NRAS and BCL2) (Table S1 and Figs. S2, S7-S9 in Supporting information) [27]. We could find that intramolecular parallel (DNA G4s: c-myc, myc 22 and c-kit1; RNA G4s: NRAS and BCL2) and hybrid (H24, bcl-2 2345 and M24) G4s could all disassemble H-aggregates of TC-P4 to dimer and monomer accompanying with the enhancement of fluorescence intensity of TC-P4. TC-P4 is negative to intermolecular parallel G4 (H7) and antiparallel G4s (A22, A24, and TBA). H7 has a covered terminal G-tetrad, and A22, A24, and TBA have diagonal loops or two lateral loops opposite to each other at the terminal G-tetrads. These structural properties enhanced the steric hindrance of G4s interacting with TC-P4 by a clip-like binding mode. TBA with one exposed terminal G-tetrad and two lateral loops opposite to each other on the other hand, might be the key reason why the weak binding affinity of TC-P4 to TBA, suggesting that TC-P4 could not bind to G4s with one covered G-tetrad. TC-P4 could recognize intramolecular parallel G4s (including DNA and RNA G4s) and hybrid G4s from the antiparallel G4s, duplex and single-stranded DNAs.
In summary, the spectral characteristics of TC and TC-P4 in solution were investigated. In PBS(K+), TC forms mixture aggregates and TC-P4 assemble mainly H-aggregates. These aggregates could be transformed into dimer and/or monomer by different DNA templates. Thus, TC and TC-P4 could recognize c-myc DNA quadruplex from duplex and single-stranded DNAs in solution, even with naked eyes. TC-P4 showed more specific recognition signal for c-myc from duplex and single-stranded DNA templates than that of TC using UV–vis absorption spectral method. TC-P4 could also discriminate the intramolecular parallel G4s and hybrid G4s from antiparallel G4s using UV–vis absorption spectral method. This transformation of aggregate property could be used as a recognizing probe for specific DNA templates.
AcknowledgmentsThis study was financially supported by General Program of the National Natural Science Foundation of China (Nos. 21472197, 21675126 and 21778058) and "Science and Technology Service Network Initiative" of the Chinese Academy of Sciences.
Appendix A. Supplementary dataSupplementary material related to this article can be found, inthe online version, at doi:https://doi.org/10.1016/j.cclet.2018.10.011.
| [1] |
A. Lohr, M. Lysetska, F. Würthner, Angew. Chem. Int. Ed. 44 (2005) 5071-5074. |
| [2] |
L.X. Wang, J.F. Xiang, H.X. Sun, et al., Dye Pigm. 122 (2015) 382-388. DOI:10.1016/j.dyepig.2015.07.018 |
| [3] |
D. Wu, Y. Shen, J. Chen, et al., Chin. Chem. Lett. 28 (2017) 1979-1982. DOI:10.1016/j.cclet.2017.07.004 |
| [4] |
J.L. Seifert, R.E. Connor, S.A. Kushon, et al., J. Am. Chem. Soc. 121 (1999) 2987-2995. DOI:10.1021/ja984279j |
| [5] |
P.H. Doan, D.R.G. Pitter, A. Kocher, et al., J. Am. Chem. Soc. 137 (2015) 9198-9201. DOI:10.1021/jacs.5b02674 |
| [6] |
L.J. Yu, W. Gai, Q.F. Yang, et al., Chin. Chem. Lett. 26 (2015) 705-708. DOI:10.1016/j.cclet.2015.02.002 |
| [7] |
Y. Xu, H. Sugiyama, Nucleic Acids Res. 34 (2006) 949-954. DOI:10.1093/nar/gkj485 |
| [8] |
J.R. Williamson, M.K. Raghuraman, T.R. Cech, Cell 59 (1989) 871-880. DOI:10.1016/0092-8674(89)90610-7 |
| [9] |
S. Burge, G.N. Parkinson, P. Hazel, et al., Nucleic Acids Res. 34 (2006) 5402-5415. DOI:10.1093/nar/gkl655 |
| [10] |
H.J. Lipps, D. Rhodes, Trends Cell Biol. 19 (2009) 414-422. DOI:10.1016/j.tcb.2009.05.002 |
| [11] |
Y.H. Shi, H.X. Sun, J.F. Xiang, et al., Chem. Commun. 52 (2016) 7302-7305. DOI:10.1039/C6CC02930B |
| [12] |
H.B. Chen, X.F. Zhang, H.X. Sun, et al., Analyst 140 (2015) 7170-7174. DOI:10.1039/C5AN01507C |
| [13] |
Y.P. Kumar, S. Bhowmik, Das Rabindra N., Chem. Eur. J. 19 (2013) 11502-11506. DOI:10.1002/chem.v19.35 |
| [14] |
K. Iida, S. Majima, T. Nakamura, et al., Molecules 18 (2013) 4328-4341. DOI:10.3390/molecules18044328 |
| [15] |
L.J. Yu, Q.F. Yang, J.F. Xiang, et al., Anal. Methods 7 (2015) 5483-5489. DOI:10.1039/C5AY01025J |
| [16] |
L.J. Yu, Q.F. Yang, J.F. Xiang, et al., Analyst 140 (2015) 1637-1646. DOI:10.1039/C4AN01912A |
| [17] |
S. Gadde, Batchelor Elizabeth K., Kaifer Angel E., et al., Chem.-Eur. J. 15 (2009) 6025-6031. DOI:10.1002/chem.v15:24 |
| [18] |
Q.F. Yang, J.F. Xiang, S. Yang, et al., Chem. Commun. (2009) 1103-1105.
|
| [19] |
M.J. Ruedas-Rama, A. Orte, M.C. Martin-Domingo, et al., J. Phys. Chem. B 118 (2014) 6098-6106. DOI:10.1021/jp5022888 |
| [20] |
J.S. Kim, R. Kodagahally, L. Strekowski, et al., Talanta 67 (2005) 947-954. DOI:10.1016/j.talanta.2005.04.025 |
| [21] |
H. Heiko, Curr. Cancer Drug Targets 3 (2003) 163-175. DOI:10.2174/1568009033481949 |
| [22] |
K. Bergmann, O'Konski C.T., J. Phys. Chem. 67 (1963) 2169-2177. DOI:10.1021/j100804a048 |
| [23] |
N. Maizels, Nat. Struct. Mol. Biol. 13 (2006) 1055-1059. DOI:10.1038/nsmb1171 |
| [24] |
D.L. Ma, Z. Zhang, M. Wang, et al., Chem. Biol. 22 (2015) 812-828. DOI:10.1016/j.chembiol.2015.06.016 |
| [25] |
H. Cai, C. Zhou, Q. Yang, et al., Chin. Chem. Lett. 29 (2018) 531-534. DOI:10.1016/j.cclet.2017.09.010 |
| [26] |
B. Jin, X. Zhang, W. Zheng, et al., Anal. Chem. 86 (2014) 943-952. DOI:10.1021/ac403676x |
| [27] |
R. Shahid, A. Bugaut, S. Balasubramanian, Biochem 49 (2010) 8300-8306. DOI:10.1021/bi100957h |
 2019, Vol. 30
2019, Vol. 30 

