b CAS Key Laboratory of Separation Sciences for Analytical Chemistry, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China;
c University of Chinese Academy of Sciences, Beijing 100049, China
The selective cell adhesion on different substrates is crucial for the tumor cell isolation [1]. Generally, the adhesion process is regulated by the surface topography and chemical function of the substrates [2, 3]. In terms of surface topography, various microscale structures (micropillars, micropits, microtips, etc.), nanoscale structures (nanowires, nanofibers, nano-dots, etc.) and hierarchical structures consisting of both micro- and nanostructured patterns, have been constructed on the substrates to promote the adhesion of tumor cells [4-6]. Topological optimization is a key strategy to improve cell adhesion behavior. Research showed that the structures matching with the cell conformation facilitated cell adhesion [4]. However, to date, the micro- and nanomanufacturing methods are not tailor-made, resulting in weak adhesion and lack of selectivity towards the target cells. By contrast, using target cells as the templates, cell imprinting can provide the topographical sites that precisely match the surface features of the template cells, which is beneficial to improve capture efficiency and capture selectivity [7]. Additionally, cell imprinting is more versatile and can be customized according to the nature of the target cells in the absence of any micro- and nanomanufacturing equipment.
Recently, cell imprinting has been successfully developed for the isolation of microorganisms [8–10]. However, due to the fragility and deformability of mammalian cells, conventional imprinting methods cannot maintain cell integrity and morphology during imprinting process, resulting in their seldom being used to isolate tumor cells [11,12]. Moreover, the selective interaction between the imprinted sites and target cells is needed to be improved.
In terms of chemical-based cues, to enhance selective adhesion of tumor cells, many specific recognition molecules such as antibodies, peptides and aptamers are functionalized on the substrates [13-15]. Among them, the aptamers are oligonucleotide sequences with specific recognition ability to target cells. Compared with antibodies and peptides, aptamers are easy to synthesize in vitro, with good reproducibility and high stability [16]. Therefore, it is expected to achieve the high selectivity isolation of tumor cells by introducing the aptamers on the substrates with imprinted sites.
Herein, based on cell imprinting and chemical recognition, we developed a cell-patterned substrate with aptamer functionalization. As shown in Scheme 1, firstly, in order to prevent tumor cells from deforming and disintegrating during imprinting, the SMMC- 7721 cells were fixed with paraformaldehyde. Secondly, polydimethylsiloxane (PDMS) was cured on the fixed SMMC-7721 cells. After the removal of the cell templates, the cell-imprinted substrate (CIS) was prepared with imprinted sites that were precisely matched to the cell surface features. The thiol groups were then introduced on the CIS by using the silanization modification (SH-CIS). The aptamers to SMMC-7721 cells were further functionalized on the SH-CIS by click chemistry. Finally, the cell-imprinted PDMS with aptamer functionalization (APT-CIS), with the synergistic effect of the cell imprinting and the chemical affinity, was further used to regulate selective cell adhesion (the detailed experimental process was shown in Supporting information).

|
Download:
|
| Scheme 1. Fabrication of the cell-imprinted polydimethylsiloxane with aptamer functionalization (APT-CIS) for the capture and release of SMMC-7721 cells. | |
The topographical structure of the substrate surface has a very important influence on cell adhesion. The morphology of the cell imprinting sites on the CIS was derived from template cells, consisting of microscale and nanoscale structures. Optical microscopy and atomic force microscopy (AFM) were used to characterize the topology of these sites. As shown in Fig. 1a, no obvious structure was formed on the non-imprinting substrate (NIS) without aptamer due to the absence of the cell templates during cell imprinting. As shown in Figs. 1b and f, the AFM results showed that the NIS is smooth and the maximum height difference of the cross section is only 19.4 nm at the dashed line. In contrast, as shown in Fig. 1c, a large number of micro and nanostructures similarly with the target cells were distributed on the CIS due to the imprinting process. As shown in Figs. 1d, e and f, not only the morphological characteristics and topological structures of the template cells were accurately preserved on the CIS, but also the nanostructures such as pseudopods of some cells are well retained. The maximum height difference of the cross section could reach 1.32 mm at dashed line. The above results indicated that the hierarchical structures matching the target cells was successfully introduced on the substrate by using the cell imprinting.

|
Download:
|
| Fig. 1. Micrographs of the surface morphology of NIS (a) and CIS (c) after removing the cell templates. AFM images of the hierarchical structures of NIS (b) and CIS (d, e). The profile (f) showing a single imprinted site at the dashed lines of the NIS and CIS. Scale bars represent 100 mm. | |
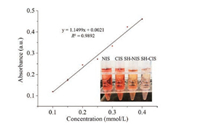
In addition to the topological structures, the chemical affinity to target cells is also important for the selective adhesion. The ZY sls aptamers can selectively bind to the SMMC-7721 cells and is easy to modification (the sequence of the ZY sls aptamers was shown in Supporting information), so the aptamers were functionalized on CIS to enhance the adhesion selectivity. In order to achieve the aptamer functionalization, thiol groups were first modified on the CIS [17]. As the control group, the NIS was also modified with thiol groups (SH-NIS). As shown in Fig. 2, the amount of the thiol groups on the substrates was measured by an iodometric method (Table S1 in Supporting information). Since there were no thiol groups on the CIS and NIS, the absorbance changes were attributed to the non-specific adsorption. After thiol modification, the color of the iodine solution became lighter due to the covalent bonding between iodine and thiol groups. The thiol groups bound to the CIS and the NIS reached 0.66 and 0.41 mmol/cm2. The above results indicated that the thiol groups were successfully modified on the substrates, which provided sufficient chemical sites for aptamer functionalization.
|
Download:
|
| Fig. 2. The calibration curve of iodometry and the images of the iodine solution after reacting with NIS, CIS, SH-NIS and SH-CIS (insert). (Error bars represent standard deviations, n = 3). | |
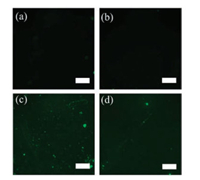
Finally, the acrydite and FITC modified aptamers were functionalized on the CIS by the thiol-ene click chemistry. To maximize the grafting of the aptamers, SH-CIS were reacted with different volumes (10–40 mL) of the aptamer solution (25 mmol/L) (Fig. S1 in Supporting information). As shown in Figs. 3a and b, the aptamers could not be modified on NIS and CIS due to the lack of the thiol groups, resulting in no fluorescent signal. In contrast, as shown in Figs. 3c and d, the fluorescent signals appeared due to the abundant thiol groups on the SH-NIS and SH-CIS, indicating that the aptamers were successfully modified on their surface to form APT-CIS and APT-NIS.
|
Download:
|
| Fig. 3. Dark field micrographs of CIS (a), NIS (b), APT-CIS (c) APT-NIS (d). Scale bars represent 100 mm. | |
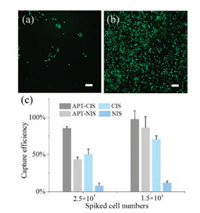
Based on the hierarchical structures, we investigate the effect of different substrates on cell adhesion performance by incubating 2.5 ×105 SMMC-7721 cells on the substrates. The incubation time was investigated as shown in Fig. S2 (Supporting information). The optimal incubation time was 3 h. As shown in Fig. 4a, only a few target cells were adsorbed on the smooth NIS, while many more cells were captured on the CIS (Fig. 4b). These results indicated that tumor cells preferred to interact with the topographical sites that precisely match the surface features of the template cells. In addition to the capture effect engineered by cell imprinting, the chemical affinity was also introduced by the aptamer functionalization. The capture efficiency was calculated to quantify the capture ability of NIS, CIS, APT-CIS and APT-NIS. As shown in Fig. 4c, due to the lack of the binding structures and affinity, NIS showed the lowest capture efficiency. The capture efficiency of CIS and APTNIS was similar, which indicated that cell imprinting and aptamer affinity are equally important for the recognition of SMMC-7721 cells. Therefore, with the synergistic effect, APT-CIS showed the highest capture efficiency for SMMC-7721 cells and could reach to 97.5% ± 11.2%, when different cell numbers were incubated on the substrates. Moreover, as shown in Fig. 5, the captured cells could be released by the complementary sequences of the aptamers with a high release efficiency up to 93.9% ± 0.8% (n = 3) which could be expected to be used for the downstream analysis.
|
Download:
|
| Fig. 4. Micrographs of SMMC-7721 cells captured on NIS (a), APT-CIS (b) and capture efficiency (c) of SMMC-7721 on NIS, CIS, APT-NIS and APT-CIS incubated with 2.5×105 and 1.5 ×104 cells. Scale bars represent 100 mm. | |

|
Download:
|
| Fig. 5. Dark field micrographs of SMMC-7721 cells before (a) and after (b) release with the complementary sequences of the ZY sls aptamer. Scale bars represent 100 mm. | |
To evaluate the capture selectivity for the target cells, we mixed SMMC-7721 with Jurkat cells at different ratios and incubated the cell mixtures on the substrates. As shown in Fig. 6, CIS had a capture efficiency from 43.0% to 72% for SMMC-7721 cells, while it is not effective, only 4% to 14%, for Jurkat cells. The enhanced contact-induced target cell adhesion could be contributed by the hierarchical structures matched precisely with the target cells, which indicated that the hierarchical structure had played a selective role in the capture of target cells. In addition, APT-NIS had a capture efficiency from 32.0% to 68% for SMMC-7721 cells, and only 6% to 9% for Jurkat cells. This indicated that the aptamer affinity had also important for the selectivity. Therefore, APT-CIS showed the highest capture efficiency and selectivity for the SMMC-7721 cells, which demonstrated that the synergistic effect of the cell imprinting and chemical affinity could improve the selective capture performance.

|
Download:
|
| Fig. 6. Capture efficiency of NIS, CIS, APT-NIS and APT-CIS for SMMC-7721 cells mixed with Jurkat cells at 1:1 (a) and 1:10 (b). | |
In summary, we prepared a cell-patterned substrate with aptamer functionalization based on cell imprinting and click chemistry. Due to the synergistic effect of topographical matching and the chemical affinity between the aptamers and target cells, the APT-CIS had high capture efficiency and selectivity to SMMC- 7721 cells. Thus, the APT-CIS holds promise in selective cell isolation fields such as the isolation of the circulating tumor cells.
AcknowledgmentsWe gratefully acknowledge the financial support from the National Key Research and Development Program (No. 2016YFA0501401), National Natural Science Foundation of China (Nos. 21575143 and 91543201), and CAS Key Project in Frontier Science (No. QYZDY-SSW-SLH017). Kaiguang Yang is the member of Youth Innovation Promotion Association, CAS (No. 2017222).
Appendix A. Supplementary dataSupplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.cclet.2018.11.006.
| [1] |
B.M. Gumbiner, Cell 84 (1996) 345-357. DOI:10.1016/S0092-8674(00)81279-9 |
| [2] |
S. Jiang, W. Wang, Y. Ding, Q. Yu, L. Yao, Chin. Chem. Lett. 29 (2018) 390-394. DOI:10.1016/j.cclet.2018.01.006 |
| [3] |
L. Liu, K. Yang, L. Zhang, Y. Zhang, Sci. Bull. 61 (2016) 1890-1891. DOI:10.1007/s11434-016-1207-7 |
| [4] |
C.S. Chen, Science 276 (1997) 1425-1428. DOI:10.1126/science.276.5317.1425 |
| [5] |
W. Wang, H. Cui, P. Zhang, et al., ACS Appl. Mater. Interface 9 (2017) 10537-10543. DOI:10.1021/acsami.7b01147 |
| [6] |
Z.X. Huang, Q. Xie, Q.P. Guo, et al., Chin. Chem. Lett. 28 (2017) 1252-1257. DOI:10.1016/j.cclet.2017.01.002 |
| [7] |
A.L. Bole, P. Manesiotis, Adv. Mater. 28 (2016) 5349-5366. DOI:10.1002/adma.v28.27 |
| [8] |
K. Eersels, van Grinsven B., M. Khorshid, et al., Langmuir 31 (2015) 2043-2050. DOI:10.1021/la5046173 |
| [9] |
K.J. Jetzschmann, G. Jágerszki, D. Dechtrirat, et al., Adv. Funct. Mater. 25 (2015) 5178-5183. DOI:10.1002/adfm.v25.32 |
| [10] |
A. Cumbo, B. Lorber, P.F. Corvini, W. Meier, P. Shahgaldian, Nat. Commun. 4 (2013) 1503. DOI:10.1038/ncomms2529 |
| [11] |
S.W. Lv, Y. Liu, M. Xie, et al., ACS Nano 10 (2016) 6201-6210. DOI:10.1021/acsnano.6b02208 |
| [12] |
O. Mashinchian, S. Bonakdar, H. Taghinejad, et al., ACS Appl. Mater. Interface 6 (2014) 13280-13292. DOI:10.1021/am503045b |
| [13] |
L. Yang, H. Sun, W. Jiang, et al., Chem. Mater. 30 (2018) 4372-4382. DOI:10.1021/acs.chemmater.8b01646 |
| [14] |
H.M. Zhang, L.J. Zhou, Z. Zhu, C.Y. Yang, Chem. -Eur. J. 22 (2016) 9886-9900. DOI:10.1002/chem.201503543 |
| [15] |
M. Labib, B. Green, R.M. Mohamadi, et al., J. Am. Chem. Soc. 138 (2016) 2476-2479. DOI:10.1021/jacs.5b10939 |
| [16] |
M. Wang, D. Yue, Q. Qiao, et al., Chin. Chem. Lett. 29 (2018) 703-706. DOI:10.1016/j.cclet.2018.03.025 |
| [17] |
J. Zhang, Y. Chen, M.A. Brook, Langmuir 29 (2013) 12432-12442. DOI:10.1021/la403425d |
 2019, Vol. 30
2019, Vol. 30 

