b University of Chinese Academy of Sciences, Beijing 100039, China
Bacteria are ubiquitous and closely relevant to human health. Illness resulting from the contaminated air, food, and water by pathogenic bacteria is a continuous serious public health problem in the world. Bacterial infection is one of the major causes of human disease and death in the world. Approximately one-third of global deaths and millions of infectious cases caused by pathogenic bacteria happen every year all over the world [1]. A thorny challenge for the treatment of infectious diseases, subject to overuse of antibiotics, is the emergence of antibiotics resistance and the developed multi-drug resistant (MDR) bacterial strains [2]. Especially to nosocomial infection, the prevalence of MDR causes some infections much more difficult to treat. What more serious is that antibiotic resistance is spreading faster than the introduction of new antibiotics nowadays, causing bacterial infection, a significant public health crisis [3]. Therefore, effective methods for rapid and efficient identification and imaging of bacteria are meaningful, and urgently required to help health professionals administer targeted treatments with reliable therapeutic.
Compared to conventional methods including culturing, gene sequencing identification techniques, surface-enchanced Raman spectroscopy (SERS) [4], and mass spectrometry (MS) [5], fluorescence analysis is rapid, easy-to-use, and highly sensitive. Various fluorescent probes for bacteria have been developed with specific receptors like antibody, aptamer, or primers to recognize biomarkers of targeted bacteria type, such as unique enzymes, mannose receptors or N-acetylglucosamine and N-acetylneuraminic acid residues on cell surfaces [6]. Recently, fluorescent sensor arrays (FSA) have also been developed to discriminate bacteria and even unknown species with rapid, easy-to-use, and highly sensitive characteristics [7].
Fluorescent probes depend on fluorescence spectra to identify and recognize bacteria species. Just as important is, depending on fluorescence microscopy [8], more dynamic information can be obtained by fluorescence imaging of bacteria [9], For example, Kasper et al. fluorescently labeled the anaerobe Bacteroides fragilis and imaged its passage through the mouse intestine in vivo and ex vivo [10]. A cyclooctyne-containing fluorophore was labeled to bacterial surfaces with azide-modified sugars via bio-orthogonal click chemistry. The probe helped to quantify total B. fragilis in the murine intestinal tract by imaging a whole live animal over time. They also obtained specific locations for the bacteria by imaging intestinal organs ex vivo. In another work, Bertozzi et al. reported a series of fluorogenic azido Si-rhodamine probes for the no-wash visualization of bacterial peptidoglycan (PG) using biocompatible copper-free click chemistry [11]. Jaffrey et al. developed genetically encode spinach-based sensors for fluorescence imaging of intracellular metabolites and proteins in living bacteria [12]. However, these reports used genetic coding technology and bioorthogonal click chemistry to locate small molecular probes in bacterial surfaces or inside bacteria, which cannot be used in intact bacterial cells.
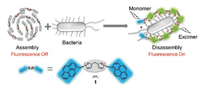
In this paper, we reported a fluorogenic probe 1 for bacteria imaging without genetic coding technology and bioorthogonal click chemistry (Fig. 1). Probe 1 was an imidazolium-derived pyrene compound, which self-assembled to form nano-particles and the pyrene fluorescence was quenched by the aggregation effects. Imidazolium acted as binding site to recognize anionic bacteria surfaces through electrostatic interaction [13]. Pyrene is a classic fluorophore that emit unique fluorescence transformed between pyrene monomer and excimer emissions [14]. When the self-assembly nano-particles interacted with anionic bacteria surfaces, synergistic effects of electrostatic interaction and hydrophobic force caused competing binding between bacteria surfaces and imidazoliums. This binding resulted in the disassembly of the aggregates to give fluorescence turn-on signal. Meanwhile, 1 bound bacteria surfaces and displayed both pyrene-excimer and pyrene-monomer fluorescence, which gave ratiometric signal. Subsequently, based on output signals of these two channels (fluorescence increase and ratiometric change), a two-dimensional analysis map could be established for bacteria identification by visual interpretation. In addition, fluorescent labeling of bacteria by 1 enabled the two-photo ratiometric imaging.
|
Download:
|
| Fig. 1. Chemical structure of 1 and its working principle as self-assembly fluorogenic probe for bacteria imaging. | |
The self-assembly property of 1 was investigated by fluorescent detection in DMSO/H2O mixtures (Fig. 2a). Probe 1 emitted strong pyrene-monomer fluorescence centered at 375 nm in DMSO solution, which indicated the dispersibility of the probe in polar solvents. When water was added into DMSO solutions, the fluorescent intensity was decreased gradually. Up to 99.5% water content, the emission intensity is even hardly discerned. It was worth noting that the intensity ratios of emission at 482 nm to that at 375 nm (I482/I375) increased rapidly with the water contents over 80% (inset in Fig. 2a). These results indicated the formation of pyrene-pyrene stacking by the self-assembly driving force. The addition of anionic surfactant SDS to the aqueous solution of 1 induced significant turn-on excimer emission (Fig. 2b). The stronger electrostatic interaction between imidazolium and SDS disassembled the aggregate perfectly, and the hydrophilic environment caused by electrostatic attraction made hydrophobic pyrene fluorophores pair-stacking together.

|
Download:
|
| Fig. 2. (a) Fluorescence spectra of 1 in DMSO/H2O mixtures (λex = 345 nm) at room temperature. (b) Fluorescence emission spectra of 1 before and after the addition of anionic surfactant SDS (20 mmol/L) in PBS buffer (10 mmol/L, pH 7.2) (λex = 345 nm). | |
Since the self-assembly probe showed high sensitivity to anionic surfactants, we felt curious whether they were also sensitive to bacteria, since the cell walls of all bacteria are nearly negatively charged. To verify the sensitivity of the probe, six different species of bacterial suspensions were separately added to the aqueous solution of the probes. As shown in Fig. 3a, the probe displayed turn-on fluorescence to these bacteria. Moreover, differential emission spectra including both pyrene excimer (center at 482 nm) and monomer (center at 375 nm) fluorescence have been presented for different bacteria. The bacteria may disassemble the aggregate to form pyrene-pyrene stacking. A twodimensional map with plots depending on both changes of fluorescence intensity (△S/S0) and emission ratios (I482/I375) was also established for bacteria identification as shown in Fig. 3b. All the other bacteria were successfully discriminated by visually observation. Surprisingly, besides the species, the probe can also identify the gram properties of the bacteria (Fig. 3B). We noted that gram-negative bacteria have a stronger tendency to induce pyrene monomer emissions. It must attribute to the significant difference of the cell walls between gram-negative and gram-positive bacteria.

|
Download:
|
| Fig. 3. (a) Fluorescence spectra of the probe (10 μmol/L) treated with six species of bacteria (OD600 = 0.5) and other ions (ATP, PPi, PO43-, CO32-, Br-, 400 μmol/L) (λex = 345 nm). (b) Two-dimensional plots from fluorescent increment (△S/S0) versus fluorescent emission ratio (I482/I375). Each circular region represents one single bacterial species, and each species have five replicates. | |
The fluorescence detection of the probe with various anionic species was conducted to examine the selectivity for bacteria. Imidazolium has been reported to have a stronger binding with phosphate ions over other anions [15]. For example, several imidazolium-derived fluorescent probes had been developed to recognize ATP/ADP, pyrophosphate and phosphate [16-18]. All these small phosphate-containing anions are of biological importance, and widely distributed in the biological system with high concentrations. We then added these anions at high concentration (400 μmol/L) to the aqueous solution of the probe, respectively. As shown in Fig. 3a, highly concentrated phosphate-containing anions, such as ATP, cannot disassemble the probe, which allows the probe to label bacteria without the interference with environmental factors.
We then sought to examine the imaging properties of the probe for bacteria (Fig. 4). E. coli were incubated with 10 μmol/L 1 for 20 min. The bacteria disassembled the aggregate and the probe labeled the bacteria surfaces. A high turn-on fluorescence response can be observed (Fig. 4e). The fluorescence turn-on response makes our probe a good fluorogenic dye for bacteria staining. More importantly, it does not require wash out the unreacted or nonspecifically bound probes before imaging. As shown in Fig. 4f, fluorescence was only observed from the stained bacteria.

|
Download:
|
| Fig. 4. Fluorescence imaging of E. coli (a–c) and E. coli incubated with 10 μmol/L of 1 (d–f) for 20 min and visualized under bright field. | |
In conclusion, we have reported a fluorogenic probe for bacteria imaging. The probe 1 was an imidazolium-derived pyrene compound, which self-assembled to form nano-particles and the pyrene fluorescence was quenched by the aggregation effects. This imidazolium-derived pyrene aggregate bound anionic bacteria surface, and disassembled to form various combinations of pyrene monomer and excimer binding modes, as controlled by the differential interaction with various bacteria surfaces. Depending on this transformable ability, our probe can rapidly identify 6 species of bacteria. The probe bound bacteria surfaces and displayed both pyrene-excimer and pyrene-monomer fluorescence, which gave ratiometric signal. Then, fluorescent labeling by the probe enabled the two-photo ratiometric imaging of bacteria. The ability of discriminating bacterial surface differences makes it possible for our one-molecule sensor to detect even more bacterial species than our current report. We are currently using this sensor to study more bacterial species and track bacterial resistance.
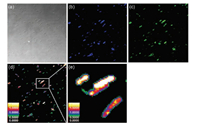
Remarkably, the probe may be able to provide quantitative information of potential distribution on bacterial surfaces, owing to its competence of performing ratiometric fluorescent imaging (Fig. 5). The cell walls of all bacteria are neatly negatively charged, although the surface structures of gram-negative and grampositive bacteria are significantly different. In general, gramnegative bacteria surface is made up of two layers of plasma membrane. The outer membrane carries amounts of lipopolysaccharide (LPS) with negative charges. gram-positive bacteria contain a single cell membrane surrounded by a thick layer of peptidoglycan, which is threaded through with abundent negatively charged teichuronic acid and lipoteichoic acids. The response of the probe to various bacteria displayed with various fluorescence profiles, which should be due to the different distribution of anions on the bacterial surfaces. Depending on these fluorescence profiles, we can discriminate different species of bacteria (Fig. 3b). Depending on two-photon microscopy (TPM), the anion distribution on bacterial surfaces was possibly mapped via ratiometric imaging.
|
Download:
|
| Fig. 5. Two-photon ratiometric confocal microscopy images of E. coli treated with 1. (a) Bright fields of E. coli; (b) Fluorescence imaging of E. coli from blue channel, collected at 420–460 nm and excited at 780 nm; (c) Fluorescence imaging of E. coli from green channel, collected at 475–540 nm and excited at 780 nm; (d) The ratiometric images (IGreen/ IBlue) were obtained by mediating the green channel image with the related blue channel image by using Image J software; (e) Higher magnification of white boxed area in (d). | |
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (Nos. 21878286, 21502189), DICP (Nos. DMTO201603, TMSR201601).
Appendix A. Supplementary dataSupplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j. cclet.2018.11.031.
| [1] |
K.E. Jones, N.G. Patel, M.A. Levy, et al., Nature 451 (2008) 990-993. DOI:10.1038/nature06536 |
| [2] |
P.C. Ray, S.A. Khan, A.K. Singh, et al., Chem. Soc. Rev. 41 (2012) 3193-3209. DOI:10.1039/c2cs15340h |
| [3] |
D.J. Payne, M.N. Gwynn, D.J. Holmes, et al., Nat. Rev. Drug Discov. 6 (2007) 29-40. DOI:10.1038/nrd2201 |
| [4] |
R. Chauvet, F. Lagarde, T. Charrier, et al., Appl. Spectrosc. Rew. 52 (2017) 123-144. DOI:10.1080/05704928.2016.1209760 |
| [5] |
M. Welker, Proteomics 11 (2011) 3143-3153. DOI:10.1002/pmic.201100049 |
| [6] |
(a) J.D. Wang, X.H. Wang, Y. Li, et al., Anal. Sci. 28 (2012) 237-241; (b) S.M. Hossain, C. Ozimok, C. Sicard, et al., Anal. Bioanal. Chem. 403 (2012) 1567-1576. |
| [7] |
(a) R.L. Phillips, O.R. Miranda, C.C. You, et al., Angew. Chem. Int. Ed. 47 (2008) 590-2594; (b) J. Han, H. Cheng, B. Wang, et al., Angew. Chem. Int. Ed. 56 (2017) 15246-15251; (c) W. Chen, Q. Li, W. Zheng, et al., Angew. Chem. Int. Ed. 53 (2014) 13734-13739. |
| [8] |
(a) S. Leng, Q. Qiao, Y. Gao, et al., Chin. Chem. Lett. 28 (2017) 1911-1915; (b) L. Peng, Y. Xu, P. Zou, Chin. Chem. Lett. 28 (2017) 1925-1928; (c) Z. Xu, J. Chen, L. Hu, et al., Chin. Chem. Lett. 28 (2017) 1935-1942; (d) P. Ning, W. Wang, M. Chen, et al., Chin. Chem. Lett. 28 (2017) 1943-1951; (e) D. Wu, Y. Shen, J. Chen, et al., Chin. Chem. Lett. 28 (2017) 1979-1982. |
| [9] |
(a) E. Zhao, Y. Chen, S. Chen, et al., Adv. Mater. 27 (2015) 4931-4937; (b) T. Gao, X. Cao, J. Dong, et al., Dyes Pigm. 143 (2017) 436-443; (c) Y. Li, X. Liu, X. Yang, et al., ACS Nano 11 (2017) 10672-10680. |
| [10] |
N. Geva-Zatorsky, D. Alvarez, J.E. Hudak, et al., Nat. Med. 21 (2015) 1091-1100. DOI:10.1038/nm.3929 |
| [11] |
S. Peyton, M.S. Siegrist, A.J. Cullen, et al., Proc. Natl. Acad. Sci. U. S. A. 111 (2014) 5456-5461. DOI:10.1073/pnas.1322727111 |
| [12] |
R.L. Strack, W. Song, S.R. Jaffrey, Nat. Protocol. 9 (2014) 146-155. DOI:10.1038/nprot.2014.001 |
| [13] |
Z. Xu, N.J. Singh, J. Lim, et al., J. Am. Chem. Soc. 131 (2009) 15528-15533. DOI:10.1021/ja906855a |
| [14] |
Z. Li, Y. Li, D. Wang, et al., Chin. Chem. Lett. 29 (2018) 1645-1647. DOI:10.1016/j.cclet.2018.01.056 |
| [15] |
Z. Xu, S.K. Kim, J. Yoon, Chem. Soc. Rev. 39 (2010) 1457-1466. DOI:10.1039/b918937h |
| [16] |
Z. Xu, N.J. Singh, J. Lim, et al., J. Am. Chem. Soc. 131 (2009) 15528-15533. DOI:10.1021/ja906855a |
| [17] |
Z. Xu, D.R. Spring, J. Yoon, Chem. Asian J. 6 (2011) 2114-2122. DOI:10.1002/asia.201100120 |
| [18] |
Z. Xu, J.Y. Choi, J. Yoon, Bull. Korean Chem. Soc. 32 (2011) 1371-1374. DOI:10.5012/bkcs.2011.32.4.1371 |
 2019, Vol. 30
2019, Vol. 30 

