b Key Laboratory of Biosynthesis of Natural Products of National Health and Family Planning Commission, Institute of Materia Medica, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100050, China
Fungi are a huge resource of natural products with novel structure and significant biological activity [1, 2]. Phenylspirodrimane dimers from fungal Stachybotrys genus structurally consist of two phenylspirodrimane monomers connected by various linkage units. They are usually formed due to the reactive property of nucleophilic benzene ring and aldehyde moiety in the phenylspirodrimane monomers, such as mer-NF5003E [3], stachybotrydial [4] and K-76 [5]. The most common linkage of the dimers is nitrogen-containing heterocycle or aliphatic chain between two phenyl groups [6-8]. Notably, phenylspirodrimane dimers with oxygen heterocycle linkage have been identified recently, including two hetero-dimers chartarolides A–B [9] fused by a 6/6 oxygen heterocycle isolated from S. chartarum WGC-25C-6, and three homo-dimers bistachybotrysins A–C [10] coupled by a 6/7 oxygen heterocycle isolated from S. chartarum CGMCC 3.5365 (China General Microbiological Culture Collection Center, CGMCC). Most recently, two new phenylspirodrimane dimers stachartone A [11] and stachartarin A [12] have been reported, however, their absolute configurations remained unsolved. To the best of our knowledge, these are all members of phenylspirodrimane dimers reported to date.
Phenylspirodrimane dimers exhibit a wide range of biological properties, including antihyperlipidemic effects in HepG2 cells [6], antibacterial activity against methicillin-resistant Staphylococcus aureus (MRSA) [7], anti-HIV-1 activity [8], and cytotoxic activities [9, 10]. In the course of our ongoing systematical investigation on more novel biologically active metabolites of S. chartarum CGMCC 3.5365, a fraction containing phenylspirodrimane dimers was targeted by analyses of HPLC-UV/MS profile, in which the molecular weights about two-fold of phenylspirodrimane monomer and identical UV spectra were observed. Further detailed isolation and purification of the target fraction led to the identification of one new pair of stereoisomeric phenylspirodrimane dimers containing a rare cyclopentanone-ring linkage, namely bistachybotrysins D and E (1 and 2, Fig. 1). Herein, we report their isolation, structural elucidation including the absolute configurations, plausible biogenetic pathway, and biological activity.

|
Download:
|
| Fig. 1. Chemical structures of compounds 1 and 2. | |
The fermentation, extraction, and isolation of the fungal strain S. chartarum CGMCC 3.5365 were performed as described previously [13, 14]. The target fraction (9.72 g) from EtOAc extract of the culture filtrate and mycelia was subjected to silica gel column chromatography (CC), MCI CC and semi-preparative HPLC to afford 1 (6.5 mg) and 2 (40.2 mg).
Bistachybotrysin D (1): White amorphous powder; [α]D25 -35.6, (c 0.54, MeOH); UV λmax (MeOH, nm) (logε): 215 (4.64), 232 (4.43), and 288 (3.94); ECD λmax (MeOH, nm) (Δε): 221 (-13.31), 238 (7.01), 289 (3.54), and 345 (-4.19); IR vmax (cm-1): 3407, 2937, 1705, 1621, 1457, 1385, 1347, 1254, 1075, and 896; 1H and 13C NMR spectroscopic data can be found in Table 1; HR-ESI-MS: m/z 773.4623 [M + H]+ (calcd. for C47H65O9: 773.4623), 795.4405 [M + Na]+.
|
|
Table 1 1H (600 MHz) and 13C (150 MHz) NMR data of compounds 1 and 2 in DMSO-d6. |
Bistachybotrysin E (2): White amorphous powder; [α]D25 -8.13, (c 0.64, MeOH); UV λmax (MeOH, nm) (logε): 214 (4.66), 233 (4.50), and 289 (4.02); ECD λmax (MeOH, nm) (Δε): 220 (17.57), 239 (-9.11), 287 (-12.31), and 343 (4.72); IR vmax (cm-1): 3382, 2934, 1703, 1621, 1458, 1387, 1349, 1255, 1037, and 896; 1H and 13C NMR spectroscopic data can be found in Table 1; HR-ESI-MS: m/z 773.4597 [M + H]+ (calcd. for C47H65O9, 773.4623), m/z 795.4407 [M + Na]+.
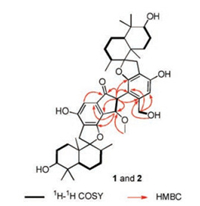
Compound 1 was obtained as white amorphous powder. Its molecular formula was determined to be C47H64O9 by the positive HR-ESI-MS ion peak at m/z 773.4623 [M + H]+ (calcd. for C47H65O9: 773.4623), corresponding to 16 degrees of unsaturation. The IR absorption bands at 3407, 1705, 1621, and 896 cm-1 indicated the presence of hydroxyl, carbonyl, and aromatic groups. By comparison with reported stachybotrysins A-G [14] and bistachybotrysins A–C [10], two sets of typical phenylspirodrimane signals were observed in the 1H NMR spectrum (Table 1), including four pairs of methyl resonances (H3-12/120, H3-13/13', H3-14/14', and H3-15/ 15'), one pair of aromatic singlet protons (H-18/18'), and one pair of oxygenated methine protons (H-3/3'). The 13C NMR data (Table 1) and DEPT (Fig. S6 in Supporting information) spectroscopic data exhibited 47 carbon resonances including 12 aromatic resonances for two phenyl groups and a carbonyl carbon, most of which appeared in pairs except for those at δC 199.6 (C-22), δC 61.3 (C-22'), δC 82.9 (C-23), δC 54.8 (C-23'), and δC 58.1 (C-1"). The above data strongly suggested a dimeric structure bearing two phenylspirodrimane units in 1. Further analysis of its 2D NMR spectroscopic data (Fig. 2) confirmed the presence of two highly identical monomers (units A and B, Fig. 1). The slight difference between units A and B is the attachment of a -CH2OH to C-19' in unit B by the 1H-1H COSY correlations of OH-22'/H-22' and HMBC correlations of H-22'/C-18', C-19', and C-20' (Fig. 2). Moreover, the unpaired signals were carefully assigned by the 1H-1H COSY coupling of H-23/H-23' and HMBC correlations (Fig. 2) of H-23'/C- 19', C-20', C-21', C-2', C-22, and C-23; H-23/C-20', C-19, C-20, C-21, and C-1"; H-18/C-22; and H-1"/C-23; which established a rare 3- (hydroxymethyl)cyclopentanone (unit C) linkage between units A and B (Fig. 1). Such a dimeric phenylspirodrimane represented a central [6,5,6]-tricyclic carbon scaffold with a novel dimerization pattern of a 3-(hydroxymethyl)cyclopentanone core fused to one phenyl unit and connected to the other by C-C bond.
|
Download:
|
| Fig. 2. 1H-1H COSY and key HMBC correlations of compounds 1 and 2. | |
The relative configuration of 1 was deduced by NOEs (Figs. S1, S11-S13 in Supporting information). The NOE correlations of H3- 15/H3-14, H3-15/H-11, H3-15/H-8, H-3/H3-14, and OH-3/H-5 in unit A, as well as H3-150/H3-140, H3-15'/H-11', H3-15'/H-8', H-3'/H3-14', and OH-3'/H-5' in unit B indicated that both units A and B of 1 possessed the same relative configuration as those of stachybotrysins A-G [14], chartarolides A-G [9], and bistachybotrysins A–C [10]. Thus, unit A and unit B were assigned to possess 3/3'R, 5/5'S, 8/8'R, 9/9'R, and 10/10'S configuration by comparison of the NMR data with those reported in literatures [9, 10, 14], as well as from biogenetic considerations. Vicinal coupling constants of JH-23/H-23' of 23, 23'-disubstituted indanone substructure of 1 are reported to be 4~5 Hz for trans-isomer and 8 Hz for cis-isomer [15-17]. Therefore, the small vicinal coupling constants of JH-23/H-23' (5.1 Hz) in 1 suggested a characteristic trans-orientation of H-23 and H-23'.
The absolute stereochemistry of 1 was deduced by ECD. The trans-orientation of H-23 and H-23' in 1 indicated the absolute configuration for C-23 and C-23' should be 23R, 23'R or 23S, 23'S. The sign of ECD spectrum for 23, 23'-disubstituted indanone substructure of 1 depends mainly on the configuration of C-23 (negative for 23R and positive for 23S), regardless of the configuration of C-23' and the solvent [18-20]. Thus, 23R-1 and 23S-1 should exhibit negative and positive Cotton effects, respectively. A negative CE at 345 nm in the ECD spectrum of 1 was observed (Fig. S15 in Supporting information), indicating R configuration for C-23, i.e., a 23R, 23'R configuration for 1. To further confirm the above absolute configuration of 1, we calculated the theoretical ECD of two stereoisomers (3R, 5S, 8R, 9R, 10S, 23R, 23'R, 3'R, 5'S, 8'R, 9'R, 10'S)-1 and (3R, 5S, 8R, 9R, 10S, 23S, 23'S, 3'R, 5'S, 8'R, 9'R, 10'S)-1 using the TD-DFT method at the B3LYP/6-31G(d) level. Comparison of the calculated ECD with the experimental ECD, the better match to the absolute configuration of 1 was (3R, 5S, 8R, 9R, 10S, 23R, 23'R, 3'R, 5'S, 8'R, 9'R, 10'S)-1, identical to the above assignment (Fig. 3). Therefore, the structure of 1 was determined and given the name of bistachybotrysin D.

|
Download:
|
| Fig. 3. The calculated ECD spectra of (23R, 23'R)-1 and (23S, 23'S)-1, and the experimental ECD spectra of compounds 1 and 2. | |
The absolute stereochemistry of 1 was deduced by ECD. The trans-orientation of H-23 and H-23' in 1 indicated the absolute configuration for C-23 and C-23' should be 23R, 23'R or 23S, 23'S. The sign of ECD spectrum for 23, 23'-disubstituted indanone substructure of 1 depends mainly on the configuration of C-23 (negative for 23R and positive for 23S), regardless of the configuration of C-23' and the solvent [18-20]. Thus, 23R-1 and 23S-1 should exhibit negative and positive Cotton effects, respectively. A negative CE at 345 nm in the ECD spectrum of 1 was observed (Fig. S15 in Supporting information), indicating R configuration for C-23, i.e., a 23R, 23'R configuration for 1. To further confirm the above absolute configuration of 1, we calculated the theoretical ECD of two stereoisomers (3R, 5S, 8R, 9R, 10S, 23R, 23'R, 3'R, 5'S, 8'R, 9'R, 10'S)-1 and (3R, 5S, 8R, 9R, 10S, 23S, 23'S, 3'R, 5'S, 8'R, 9'R, 10'S)-1 using the TD-DFT method at the B3LYP/6-31G(d) level. Comparison of the calculated ECD with the experimental ECD, the better match to the absolute configuration of 1 was (3R, 5S, 8R, 9R, 10S, 23R, 23'R, 3'R, 5'S, 8'R, 9'R, 10'S)-1, identical to the above assignment (Fig. 3). Therefore, the structure of 1 was determined and given the name of bistachybotrysin D.
Compound 2 was obtained as white amorphous powder. Its molecularformula, C47H64O9, was determined tobe the same as that of 1 by HR-ESI-MS at an m/z of 773.4597 [M + H]+ (calcd. for C47H65O9: 773.4623). The 1H and 13C NMR data of 2 were closely similar to that of 1, and further analysis of the 2D NMR data of 2 revealed that both 1 and 2 possess the same planar structure (Fig. 2).
The similar NOE correlations of units A and B for both 1 and 2 indicated that they have the same relative configuration (Fig. S1, S25-S27 in Supporting information), which was further supported by the consistent chemical shifts and coupling constants of their protons (Table 1). The small coupling constants of JH-23/H-23' (3.3 Hz) for 2 revealed that H-23 and H-23' were also transoriented. However, the ECD spectrum of 2 (Fig. S29 in Supporting information) displayed opposite CEs to that of 1, indicating that 2 is a stereoisomer of 1 regarding the stereogenic centers in unit C. The positive CE at 343 nm in the ECD spectrum of 2 (Fig. S29) indicated S configuration for C-23, and a 23S, 23'S configuration for 2, which was further supported by the good match of the experimental ECD curves of 2 to the calculated ECD of (3R, 5S, 8R, 9R, 10S, 23S, 23'S, 3'R, 5'S, 8'R, 9'R, 10'S)-1 (Fig. 3). Thus, the structure of 2 was determined, and named bistachybotrysin E.
Compounds 1 and 2 are a class of new phenylspirodrimane dimers with a rare five-membered carbon ring linkage. The central [6,5,6]-tricyclic carbon scaffold represents a novel dimerization pattern of a central cyclopentanone core fused to one phenyl unit and connected to the other one by C-C bond. The biosynthetic pathway of 1 and 2 was postulated in Scheme 1. Phenylspirodrimane monomers are assembled from orsellinic acid and farnesyldiphosphate (FPP) by prenylation, reduction and cyclization [21]. Further oxidation and hydroxylation yield stachybotrydial and mer-NF5003E, which undergo a pinacol coupling reaction [7, 22] between two aldehyde groups to generate a vicinal diol intermediate 3A. 3B is formed by dehydration of H-22 and OH- 23' of 3A. Subsequent non-stereoselective Aldol reaction of 3B forms one pair of stereoisomers, and further O-methylation affords 1 and 2, respectively.

|
Download:
|
| Scheme 1. Plausible biosynthetic pathway of 1 and 2. | |
Compounds 1 and 2 were evaluated for in vitro cytotoxicity (Table 2) against five human tumor cell lines (colorectal carcinoma HCT116, lung carcinoma NCI-H460, gastric carcinoma BGC823, medulloblastoma Daoy, and liver carcinoma HepG2) and neural anti-inflammatory activity. Compounds 1 and 2 displayed potent cytotoxic activity against the cell lines HCT116, BGC823, Daoy and HepG2 with IC50 values ranging from 6.7 μmol/L to 11.6 μmol/L. In addition, compound 1 exerted neural anti-inflammatory activity by inhibiting LPS-induced NO production in BV2 cells with the inhibitory rate of 61.1% at 10 μmol/L, which was comparable to curcumin, a positive control with the inhibitory rate of 67.6%.
|
|
Table 2 Cytotoxicity of compounds 1 and 2. |
In summary, we report one new pair of stereoisomeric phenylspirodrimane dimers bistachybotrysins D and E (1 and 2), which exhibited potent cytotoxic activity against several tumor cell lines and neural anti-inflammatory activity. Their absolute configurations were solved by calculated ECD. Biogenetically, compounds 1 and 2 contained a unique central [6,5,6]-tricyclic carbon scaffold generated from a proposed pinacol coupling reaction and Aldol reaction, indicating the diversity of dimerization pattern in phenylspirodrimane compounds family. Moreover, non-stereospecificity of the biosynthetic enzymes might contribute to the generation of diverse stereoisomeric pairs of natural products [23].
AcknowledgmentThis work was financially supported by CAMS Innovation Fund for Medical Sciences (Nos. CIFMS-2016-I2M-3-012 and CAMS-I2M- 2-002).
Appendix A. Supplementary dataSupplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.cclet.2018.04.031.
| [1] |
J.M. Liu, D.W. Zhang, M. Zhang, et al., Chin. Chem. Lett. 28 (2017) 248-252. DOI:10.1016/j.cclet.2016.07.031 |
| [2] |
D.W. Zhang, X.Y. Tao, J.M. Liu, et al., Chin. Chem. Lett. 27 (2016) 640-642. DOI:10.1016/j.cclet.2016.02.005 |
| [3] |
R. Kaneto, K. Dobashi, I. Kojima, et al., J. Antibiot. 47 (1994) 727-730. DOI:10.7164/antibiotics.47.727 |
| [4] |
T.W. Lin, W.W. Chang, C.C. Chen, Y.C. Tsai, Biochem. Bioph. Res. Co. 331 (2005) 953-957. DOI:10.1016/j.bbrc.2005.03.232 |
| [5] |
H. Kaise, M. Shinohara, W. Miyazaki, et al., J. Chem. Soc. Chem. Commun. (1979) 726-727. |
| [6] |
Y. Li, C. Wu, D. Liu, et al., J. Nat. Prod. 77 (2014) 138-147. DOI:10.1021/np400824u |
| [7] |
B. Wu, V. Oesker, J. Wiese, et al., Mar. Drugs 12 (2014) 1924-1938. DOI:10.3390/md12041924 |
| [8] |
B.E. Roggo, F. Petersen, M. Sills, et al., J. Antibiot. 49 (1996) 13-19. DOI:10.7164/antibiotics.49.13 |
| [9] |
D. Liu, Y. Li, X. Li, et al., Tetrahedron Lett. 58 (2017) 1826-1829. DOI:10.1016/j.tetlet.2017.03.079 |
| [10] |
J. Zhao, J. Feng, Z. Tan, et al., Bioorg. Med. Chem. Lett. 28 (2018) 355-359. DOI:10.1016/j.bmcl.2017.12.039 |
| [11] |
Z.G. Ding, J.Y. Zhao, J.H. Ding, et al., Nat. Prod. Res. 32 (2018) 1699-1705. DOI:10.1080/14786419.2017.1396599 |
| [12] |
Z.G. Ding, J.H. Ding, J.Y. Zhao, et al., Fitoterapia 125 (2018) 94-97. DOI:10.1016/j.fitote.2017.12.022 |
| [13] |
J. Zhao, J. Liu, Y. Shen, et al., Phytochem Lett. 20 (2017) 289-294. DOI:10.1016/j.phytol.2017.04.031 |
| [14] |
J. Zhao, J. Feng, Z. Tan, et al., J. Nat. Prod. 80 (2017) 1819-1826. DOI:10.1021/acs.jnatprod.7b00014 |
| [15] |
S. Forsén, B. Gestblom, R.A. Hoffman, S. Rodmar, J. Mol. Spectrosc. 21 (1966) 372-385. DOI:10.1016/0022-2852(66)90163-9 |
| [16] |
P.H. Lacy, D.C.C. Smith, J. Chem. Soc. Perkin. Trans. 1 (1974) 2617-2619. |
| [17] |
K.M.E. Ng, T.C. McMorris, Can. J. Chem. 62 (1984) 1945-1953. DOI:10.1139/v84-334 |
| [18] |
M. Kuroyanagi, M. Fukuoka, K. Yoshihira, S. Natori, Chem. Pharm. Bull. 27 (1979) 592-601. DOI:10.1248/cpb.27.592 |
| [19] |
M. Kuroyanagi, M. Fukuoka, K. Yoshihira, S. Natori, Chem. Pharm. Bull. 27 (1979) 731-741. DOI:10.1248/cpb.27.731 |
| [20] |
S.J. Uddin, T.L.H. Jason, K.D. Beattie, I.D. Grice, E. Tiralongo, J. Nat. Prod. 74 (2011) 2010-2013. DOI:10.1021/np2004598 |
| [21] |
C. Li, Y. Matsuda, H. Gao, et al., ChemBioChem 17 (2016) 904-907. DOI:10.1002/cbic.v17.10 |
| [22] |
K. Ogawa, M. Nakamura, M. Hayashi, et al., J. Antibiot. 48 (1995) 1396-1400. DOI:10.7164/antibiotics.48.1396 |
| [23] |
J.M. Finefield, D.H. Sherman, M. Kreitman, R.M. Williams, Angew. Chem. Int. Edit. 51 (2012) 4802-4836. DOI:10.1002/anie.v51.20 |
 2019, Vol. 30
2019, Vol. 30 



