b College of Chemistry and Chemical Engineering, Qingdao University, Qingdao 266071, China
Bacterial infections are among the most serious health problems and have led to increased morbidity, mortality and cost of health care for patients [1, 2]. In particular, infections resulting from gram-positive bacteria can cause a variety of serious diseases including sepsis, bacteremia, pneumonia, osteomyelitis and endocarditis [3, 4]. These infections can be effectively controlled by various antibiotics, but the emergence of resistant and more virulent strains of bacteria has outpaced the development of new antibiotics, e.g., penicillin, tetracycline, streptomycin and vancomycin [5]. Some metal based nanomaterials have been widely exploited as antibacterial agents due to their broad spectrum of antimicrobial activity [6].
Copper is a relatively abundant element which is highly delicate in the life process, it is essential for metalloproteins and enzymes in the living systems. At a high concentration level, copper induces an inhibition effect on bacteria growth and it exhibits toxic effect on most microorganisms. Copper nanoparticles (CuNPs) possess the capability of killing bacteria cells by damaging the cell wall/ membrane [7, 8]. In comparison with CuNPs, copper nanoclusters (CuNCs) consist of a few to a few hundred of Cu atoms with a diameter of < 5 nm. It has been reported that CuNCs can serve as a new category of luminescent nanostructure for bioimaging and light emitting devices [9]. However, a literature search indicates that there is so far no report on the study of antibacterial activity of CuNCs. Appropriate functionalization or capping of CuNCs may be a feasible approach for endowing them bactericidal capability.
Tannic acid (TA) is a natural polyphenolic molecule consisting of a central core of glucose linking to ten gallic acid groups through ester bonds, and its chemical structure is shown in Fig. S1 (Supporting information) [10]. It is well known that TA possesses antibacterial, antioxidant, anticarcinogenic and antimutagenic activities [11]. The numerous hydroxyl groups in TA configuration can interact with various species involved in the life process, e.g., proteins, digestive enzymes, carbohydrates and metal ions, which provide many intriguing physical and chemical properties of TAbased materials [12]. TA has been used as stabilizing agent to protect the nanoclusters from aggregation and oxidation which facilitates bioimaging and biosensing [13-16]. By taking into account the antibacterial activity of TA, it might be feasible and highly interesting to integrate bactericidal feature into TA stabilized CuNCs.
For the purpose of realizing the above hypothesis, we employ TA as a stabilizing agent to produce novel CuNCs (TA-CuNCs). In the TA-CuNCs, synergistic effect of TA moiety and the copper nanoclusters is observed and demonstrated, which endows TA-CuNCs significantly high bactericidal capability. TA-CuNCs are selective to inhibit the growth of gram-positive bacteria Staphylococcus aureus (S. aureus) and Bacillus subtilis (B. subtilis) against gram-negative bacteria Escherichia coli O157:H7 (E. coli) and Pseudomonas aeruginosa (P. aeruginosa). The mechanisms of selective bactericidal effect toward gram-positive bacteria are further investigated.
TEM image as shown in Fig. S2A (Supporting information) illustrates spherical TA-CuNCs which are well dispersed in aqueous medium. Histogram of the particle size distribution achieved from multiple TEM images is shown in Fig. S2B (Supporting information), demonstrating an average size of 1.0 ± 0.7 nm (200 particles were taken into account for calculation).
XPS spectra are recorded to obtain the elemental composition and oxidation state of copper in TA-CuNCs. Two intense peaks located at 932.7 eV and 952.4 eV are attributed to 2p3/2 and 2p1/2 features of Cu(0) (Fig. S2C). Moreover, no characteristic peak of Cu(Ⅱ) at 942 eV is observed, indicating the absence of oxidation (Cu2+) in TA-CuNCs. Considering that the 2p3/2 binding energy of Cu (0) is very close to that of Cu(Ⅰ) [17], the valence state of TA-CuNCs most likely to locate in between 0 and +1. C 1s spectrum (Fig. S2D) shows three peaks at 284.6, 286.3 and 288.4 eV, due to the contribution of C═C, C═O and O-C = O. O 1s spectrum (Fig. S2E) exhibits three peaks at 531.7 eV, 532.5 eV, and 533.2 eV, which are attributed to Cu–O [18], -OH/C═O, and C–O–C/C-OH groups. These observations clearly suggest the presence of TA on the surface of TA-CuNCs.
UV-vis absorption spectrum of TA-CuNCs (Fig. S2F in Supporting information) shows two distinguish peaks within the UV region at 210 nm and 274 nm, respectively. These molecular-like optical transition absorbance bands are due to the quasi continuous electronic energy band structure and quantum confinement effects of TA-CuNCs, which is significantly different from a surface plasmon resonance band at 550–600 nm of CuNPs [19]. The dependence of UV–vis absorbance of TA-CuNCs aqueous dispersion on time is shown in Fig. S3 (Supporting information). It illustrates that TA-CuNCs is stable in 45 days.
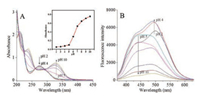
The absorption spectra of TA-CuNCs aqueous dispersion are recorded within pH 2–10 (Fig. 1A). The absorption at 274 nm is attributed to the usual n-π* transition of the functional group C═O [20]. It decreases obviously by varying pH 2–4, and a new broad absorption at 326 nm appears and significantly increases within pH 5-10. Gallic acid is a product during hydrolysis of tannic acid by ester bonds cleavage using tannase. In this work, tannase is absent and tannic acid should not be biocatalyzed to gallic acid [21]. The change of UV–vis spectra should be mainly due to protonation and deprotonation of the functional phenolic hydroxy group [22]. This phenomenon probably arises from the deprotonation of the phenolic hydroxyl and carboxyl functional groups on TA-CuNCs. The electric potential variation of the TA-CuNCs with pH is shown in Table S1 (Supporting information). The decrement of electric potential of TA-CuNCs aqueous dispersion indicates that TA-CuNCs is deprotonated when pH increased. The pH-dependent absorption behavior clearly demonstrated the presence of TA moiety on the surface of CuNCs.
|
Download:
|
| Fig. 1. (A) UV–vis absorption spectra of TA-CuNCs aqueous dispersion within pH 2–10. The inset shows pH dependent behavior of the absorbance at 326 nm; (B) Fluorescence emission spectra of TA-CuNCs aqueous dispersion at λex 350 nm within pH 2–10. | |
Fig. S2F illustrates fluorescence emission of TA-CuNCs at λex/ λem 350 nm/500 nm and yellow luminescence under UV illumination (Inset, right). pH-Induced variation of the fluorescence spectra of TA-CuNCs is illustrated in Fig. 1B, where the maximum emission wavelength shifts from 500 nm to 440 nm by varying pH 2–10. This result is obviously in accordance with pH-induced deprotonation of phenol and carboxyl functional groups of carbon dots [22]. The variation of UV–vis and fluorescence spectra is ascribed to two prototropic equilibria, i.e., phenol ↔ phenolate and carboxylic ↔ carboxylate.
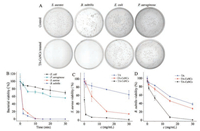
The antibacterial capability of TA-CuNCs is investigated with agar plate method. Fig. 2A indicates that TA-CuNCs dramatically inhibit the growth of gram-positive bacteria S. aureus and B. subtilis, while their antibacterial effect for gram-negative bacteria E. coli and P. aeruginosa is not obvious. Fig. 2B illustrates the viability variation of S. aureus, B. subtilis, E. coli and P. aeruginosa (1 × 107 CFU/mL) by treatment with 30 μg/mL TA-CuNCs at various sterilization time. It is obvious that a sterilization time of 2 min results in a significant decrease on the viability of gram-positive bacteria S. aureus and B. subtilis to 26.8% and 14.8%, respectively. However, those for the gram-negative bacteria E. coli and P. aeruginosa encountered only a slight drop to 90.0% and 92.3%, respectively. At a sterilization time of 10 min, the gram-positive bacteria is completely inhibited, but the viability of gram-negative bacteria E. coli and P. aeruginosa still remain at 85.0% and 72.0%, respectively. These results show that TA-CuNCs exhibit selective antibacterial activity against gram-positive bacteria.
|
Download:
|
| Fig. 2. (A) Photographs of 1 × 107 CFU/mL bacteria colonies, e.g., gram-positive bacteria S. aureus and B. subtilis, gram-negative bacteria E. coli and P. aeruginosa, grown on broth agar plates in the presence of 30 μg/mL TA-CuNCs under a sterilization time of 10 min; (B) The viability of 1 × 107 CFU/mL bacteria after treatment with 30 μg/mL of TACuNCs in various sterilization time (2, 5, 10, 15, 20 and 30 min); Antibacterial activities of TA-CuNCs, FA-CuNCs and TA (0, 0.1, 1, 5, 15 and 30 μg/mL) against S. aureus (C) and B. subtilis (D) by shaking for 10 min. | |
The structures of cell envelope, including cytoplasmic membrane and cell wall component, are different between grampositive and gram-negative bacteria. Gram-positive bacteria possess an outer membrane surrounding the cell wall, restricting diffusion of hydrophobic components through its peptidoglycan and lipopolysaccharide covering [23]. However, gram-negative bacteria have much less peptidoglycan. When TA acts on peptidoglycan of the gram-positive bacteria, the cell wall/ membrane is permeated more easily and exogenous substances can disturb the cytoplasmic membrane, and thus disrupt the proton motive force, electron flow, active transport and coagulation of cell contents [24]. In addition, gram-negative bacteria are negatively charged due to the large number of carboxylic and other groups [25]. On the contrary, however, gram-positive bacteria possess very weak negative charge, which facilitates the permeation of negatively charged free radicals, e.g., carboxylate radical (-COO-) and hydroxyl radicals (-OH), to penetrate into the cell membrane and kill the cells. A Zeta potential value of -11.2 mV is recorded for TA-CuNCs in aqueous medium. The negatively charged TA-CuNCs interact with gram-positive bacteria by electrostatic interaction, which further improves the bactericidal activity of TA-CuNCs [26-28].
The antibacterial performance of TA-CuNCs against S. aureus and B. subtilis at various concentration levels, i.e., 0.1, 1, 5, 15 and 30 μg/mL, is illustrated in Fig. S4 (Supporting information), where similar trends are observed as those demonstrated in Fig. 2. The minimum inhibitory concentration (MIC), with a definition of the lowest concentration of the antimicrobial agent to prevent visible growth of microorganism, is derived to be 5 μg/mL TA-CuNCs for S.aureus and 15 μg/mL for B. subtilis. These MIC values are comparable to that of standard antibiotic nalidixic acid [29].
The antibacterial activity of TA-CuNCs against gram-positive bacteria are further elucidated with folic acid (FA) stabilized CuNCs [30], shortly as FA-CuNCs, and bare TA as controls. The viability of S.aureus is significantly decreased to 13.9% after treated by 1.0 mg/mL TA-CuNCs with a sterilization time of 10 min (Fig. 2C). While the viability of S. aureus remains as high as 89.6% and 87.8% by treating with same concentration of FA-CuNCs and bare TA, respectively. Photographs of S. aureus and B. subtilis colonies grown on broth agar plates in the presence of 30 μg/mL of TA-CuNCs, FA-CuNCs and TA at a sterilization time of 10 min are shown in Figs. S5 and S6 (Supporting information). Compared to FA-CuNCs and TA treated bacteria, TA-CuNCs are remarkably effective in the inhibition of bacteria growth for S. aureus and B. subtilis. Figs. 2C and D illustrate that their antibacterial performance become even stronger with the increase of dosage. The above observations clearly demonstrate that TA-CuNCs have significantly superior antibacterial capability with respect to both FA-CuNCs and TA against gram-positive bacteria. The TA molecules on TA-CuNCs surface having a high local concentration may interact with the cell walls effectively and damage the cell walls, which lead to the death of bacteria [31-33].
Metal-based nanoparticles or nanoclusters can generate free metal ions in aqueous medium which interact with cell membrane directly and inhibit bacteria growth through multiple mechanisms, e.g., inhibition of electron transport chain and the regulation of bacterial metabolic processes [34]. In the present case, TA-CuNCs exhibit favorable antibacterial capability against gram-positive bacteria, and the antimicrobial mechanisms may include the following steps: (ⅰ) TA moiety in TA-CuNCs functions with peptidoglycan of the gram-positive bacteria and wherein destroys the cell membrane, as demonstrated by the above experimental results; (ⅱ) Negatively charged TA-CuNCs readily interact with gram-positive bacteria by electrostatic interactions, which improves the bactericide selectivity of TA-CuNCs; (ⅲ) The attack and permeation of TA-CuNCs into the bacteria interior through the membrane of the gram-positive bacteria cells may result in the release of cationic Cu+ or elemental Cu [33, 34], which cause damaging on the bacteria cell membrane, blocking the enzyme activity, changing the bacteria cell structure and ultimately inhibiting the growth of the bacteria [35].
If the bacteria cell membrane is damaged by TA-CuNCs, the release of cytoplasmaic constituents, e.g., DNA and RNA, can be quantified by monitoring the strong UV absorption at 260 nm. Fig. S7 (Supporting information) illustrates DNA and RNA release behaviors of the bacteria after incubation with 30 μg/mL of TACuNCs, by showing the optical density (OD) ratio of the bacteria suspensions. It is obviously to see that the OD260nm ratios of grampositive bacteria S. aureus and B. subtilis are significantly higher than those of the gram-negative bacteria E. coli and P. aeruginosa suspensions. This observation well agrees with the cell viability measurements, illustrating that the reflux of DNA and RNA correlates to the bacteria mortality. It also implies that the bacteria death is directly associated with the damage of bacteria cell membrane.
AFM images of TA-CuNCs treated bacteria cells are recorded in hydrated condition without any chemical modification, for the purpose of understanding the changes in the microstructure and physicochemical properties of the bacteria cells after treatment. For comparison, the phosphate buffer treated bacteria cells are also illustrated as control (Fig. 3). TA-CuNCs treated gram-positive bacteria cells show substantial alteration in their cellular morphology with respect to those of the control cells. As shown in Figs. 3A and B, after incubation with 30 μg/mL TA-CuNCs for 10 min, bacteria S. aureus and B. subtilis cells lost their cellular integrity. The membrane of S. aureus is lysed and large amount of cellular debris are found around the cells due to the leakage in the cell membrane. The external surface of B. subtilis, marked by arrows in the micrographs, is clearly ruptured. AFM images of the ultrastructural morphology of gram-negative bacteria E. coli and P. aeruginosa show typical mycelia and flagellum (Figs. 3C-ii and 3Dii). It is clearly seen that after TA-CuNCs treatment, the bacteria cell membranes of E. coli and P. aeruginosa maintain their integrity. The above observations provide a further visible insight into the antimicrobial mechanisms of TA-CuNCs.

|
Download:
|
| Fig. 3. AFM images of S. aureus (A), B. subtilis (B), E. coli (C) and P. aeruginosa cells (D). Control cells treated with phosphate buffer (ⅰ) and the bacteria cells treated with 30 μg/mL of TA-CuNCs for 10 min (ⅱ). | |
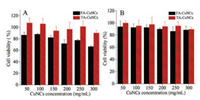
The biocompatibility of TA-CuNCs with HepG2 cells and 293 T cells is thoroughly evaluated in terms of cell proliferation, with FACuNCs as a comparison. The viability of the cells is plotted against cells is thoroughly evaluated in terms of cell proliferation, with FACuNCs as a comparison. The viability of the cells is plotted against various concentrations of TA-CuNCs and FA-CuNCs. Fig. 4A indicates virtually no cytotoxic effect of TA-CuNCs on 293 T cells. The viability of 293 T cells remains 96.7% after incubation with TACuNCs at a concentration level up to 200 μg/mL for 24 h. On the other hand, however, when incubating the cells with same concentration of FA-CuNCs, a cell viability of ca. 71.4% is achieved. Similar results are obtained with HepG2 cells as illustrated in Fig. 4B, where a cell viability of >94.2% is recorded for HepG2 cells after incubation with TA-CuNCs for 24 h. These observations clearly demonstrate the fact that TA-CuNCs induced minimum cellular toxicity. In comparison with FA-CuNCs by using folic acid (FA) as stabilizer, TA capped TA-CuNCs exhibit favorable biocompatibility
|
Download:
|
| Fig. 4. The viability of 293 T cells (A) and HepG2 cells (B) after incubating with TACuNCs and FA-CuNCs at various concentration levels (50–300 mg/mL) in 10 mmol/L PBS buffer (pH 7.4) for 24 h. | |
In the present work, we report a novel copper nanoclusters, i.e., tannic acid (TA) capped CuNCs (TA-CuNCs). There exists obvious synergistic effect between TA moiety and the nanoclusters, which endows TA-CuNCs high antibacterial activity. The bactericidal effect of TA-CuNCs is significantly higher than that of the conventional CuNCs, by taking the previously reported folic acid capped copper nanoclusters as an example. TA-CuNCs are selective to inhibit/against the growth of gram-positive bacteria by damaging the cell membrane. Meanwhile, TA-CuNCs exhibit low cytotoxicity and favorable biocompatibility, which provides a promising candidate of bactericide for infection treatment caused by gram-positive bacteria. The present approach opens up a new avenue for the development of efficient antibacterial agent via appropriate functionalization or modification of copper nanoclusters.
AcknowledgmentsThis work is financially supported by the National Natural Science Foundation of China (Nos. 21675019, 21727811, 21575020), Fundamental Research Funds for the Central Universities (Nos. N170505002, N170504017, N170507001).
Appendix A. Supplementary dataSupplementary material related to this article can befound, in the online version, at doi:https://doi.org/10.1016/j.cclet.2018.07.008.
| [1] |
M.V. Oosten, T. Schäfer, J.A.C. Gazendam, et al., Nat. Commun. 4 (2012) 2584-2591. |
| [2] |
J. Zhou, G.B. Qi, H. Wang, J. Mater. Chem. B 4 (2016) 4855-4861. |
| [3] |
H.J. Chung, T. Reiner, G. Budin, et al., ACS Nano 5 (2011) 8834-8841. DOI:10.1021/nn2029692 |
| [4] |
S.K. Choi, A. Myc, J.E. Silpe, et al., ACS Nano 7 (2013) 214-218. DOI:10.1021/nn3038995 |
| [5] |
X. Zhu, A.F. Radovic-Moreno, J. Wu, R. Langer, J.J. Shi, Nano Today 9 (2014) 478-498. DOI:10.1016/j.nantod.2014.06.003 |
| [6] |
H.A. Hemeg, Int. J. Nanomed. 12 (2017) 8211-8225. DOI:10.2147/IJN |
| [7] |
J. Ramyadevi, K. Jeyasubramanian, A. Marikani, G. Rajakumar, A.A. Rahuman, Mater. Lett. 71 (2012) 114-116. DOI:10.1016/j.matlet.2011.12.055 |
| [8] |
C.H. Deng, J.L. Gong, G.M. Zeng, et al., Chemosphere 184 (2017) 347-357. DOI:10.1016/j.chemosphere.2017.05.118 |
| [9] |
L.B. Zhang, E.K. Wang, Nano Today 9 (2014) 132-157. DOI:10.1016/j.nantod.2014.02.010 |
| [10] |
K.T. Chung, T.Y. Wong, C.I. Wei, Y.W. Huang, Y. Lin, Crit. Rev. Food Sci. 38 (1998) 421-464. DOI:10.1080/10408699891274273 |
| [11] |
M. Daglia, Curr. Opin. Biotech. 23 (2012) 174-181. DOI:10.1016/j.copbio.2011.08.007 |
| [12] |
T. Ahmad, J. Nanotechnol. 2014 (2014) 1-11. |
| [13] |
H. Cao, Z. Chen, H. Zheng, Y. Huang, Biosens. Bioelectron. 62 (2014) 189-195. DOI:10.1016/j.bios.2014.06.049 |
| [14] |
Q. Liu, Q. Lai, N. Li, X.G. Su, Microchim. Acta 185 (2018) 182-188. DOI:10.1007/s00604-017-2599-z |
| [15] |
R.S. Aparna, S.S. Syamchand, S. George, J. Clust. Sci. 28 (2017) 2223-2238. DOI:10.1007/s10876-017-1221-1 |
| [16] |
H.B. Rao, H.W. Ge, Z.W. Lu, W. Liu, Microchim. Acta 183 (2016) 1651-1657. DOI:10.1007/s00604-016-1794-7 |
| [17] |
J.S. Shen, Y.L. Chen, Q.P. Wang, et al., J. Mater. Chem. C 1 (2013) 2092-2096. DOI:10.1039/c3tc00941f |
| [18] |
J. Ma, Z.L. Zhu, B. Chen, et al., J. Mater. Chem. A 1 (2013) 4662-4666. DOI:10.1039/c3ta10329c |
| [19] |
D. Mott, J. Galkowski, L.Y. Wang, J. Luo, C.J. Zhong, Langmuir 23 (2007) 5740-5745. DOI:10.1021/la0635092 |
| [20] |
J. Luo, N. Zhang, J.P. Lai, R. Liu, X.Y. Liu, J. Hazard. Mater. 300 (2015) 615-623. DOI:10.1016/j.jhazmat.2015.07.079 |
| [21] |
B. Mahendran, N. Raman, D.J. Kim, Appl. Microbiol. Biot. 70 (2006) 444-450. DOI:10.1007/s00253-005-0082-y |
| [22] |
S.D. Choudhury, J.M. Chethodil, P.M. Gharat, P.K. Praseetha, H. Pal, J. Phys. Chem. Lett. 8 (2017) 1389-1395. DOI:10.1021/acs.jpclett.7b00153 |
| [23] |
S. Burt, Int. J. Food Microbiol. 94 (2004) 223-253. DOI:10.1016/j.ijfoodmicro.2004.03.022 |
| [24] |
F. Tian, B. Li, B.P. Ji, et al., Food Chem. 113 (2009) 173-179. DOI:10.1016/j.foodchem.2008.07.062 |
| [25] |
P. Pageni, P. Yang, Y.P. Chen, et al., Biomacromolecules 19 (2018) 417-425. DOI:10.1021/acs.biomac.7b01510 |
| [26] |
K.G. Yu, D.H. Li, C.H. Zhou, J.L. Diao, Chin. Chem. Lett. 20 (2009) 411-414. DOI:10.1016/j.cclet.2008.11.030 |
| [27] |
C. Anupama, A. Kaphle, G. Udayabhanu, Nagaraju, J. Mater. Sci.-Mater. El. 29 (2018) 4238-4249. DOI:10.1007/s10854-017-8369-1 |
| [28] |
X.D. Zhang, X.K. Chen, J.J. Yang, et al., Adv. Funct. Mater. 26 (2016) 5958-5970. DOI:10.1002/adfm.v26.33 |
| [29] |
M.S. Alam, S.M.M. Rahman, D.U. Lee, Chem. Pap. 69 (2015) 1118-1129. |
| [30] |
J.M. Xia, X. Wei, X.W. Chen, Y. Shu, J.H. Wang, Microchim. Acta 185 (2018) 205-211. DOI:10.1007/s00604-018-2743-4 |
| [31] |
J.J. Yang, X.D. Zhang, Y.H. Ma, et al., ACS Appl. Mater. Interfaces 8 (2016) 32170-32181. DOI:10.1021/acsami.6b10398 |
| [32] |
Y.W. Jiang, G. Gao, X.D. Zhang, H.R. Jia, F.G. Wu, Nanoscale 9 (2017) 15786-15795. DOI:10.1039/C7NR04679K |
| [33] |
H.Y. Wang, X.W. Hua, F.G. Wu, et al., ACS Appl. Mater. Interfaces 7 (2015) 7082-7092. DOI:10.1021/acsami.5b01214 |
| [34] |
K.Y. Zheng, M.I. Setyawati, D.T. Leong, J.P. Xie, Coordin. Chem. Rev. 357 (2018) 1-17. DOI:10.1016/j.ccr.2017.11.019 |
| [35] |
L.L. Wang, C. Hu, L.Q. Shao, Int. J. Nanomed. 12 (2017) 1227-1249. DOI:10.2147/IJN |
 2019, Vol. 30
2019, Vol. 30 

