b Shanghai Key Laboratory of Green Chemistry and Chemical Processes, School of Chemistry and Molecular Engineering, East China Normal University, Shanghai 200241, China
Plasmonic metamaterials, which possess numerous unusual optical properties such as circular dichroism (CD) [1], giant nonlinear optical activity [2], and Fano resonance [3], have drawn great attention due to their extensive applications in biosensing, imaging, photonics, and optoelectronics, etc. [4, 5]. These artificial plasmonic structures are typically constructed through either topdown [6-9] or bottom-up [10-14] approaches. However, the former ones are often restricted to resolution, fabrication, and rational three-dimensional (3D) organization of nanoarchitectures [15]. Bottom-up methods, on the other hand, provide a new pathway to overcome these challenges. Particularly, DNA with unprecedented programmability and addressability, has demonstrated to be one of the most powerful and versatile building blocks for fabrication of sophisticated architectures with well-defined shapes and functions [16-21]. In this field, DNA origami [22, 23], undoubtedly a milestone in the progress of DNA nanotechnology, has been utilized not only to construct diverse pure DNA nanostructures [24-28], but also performed as ideal templates to guide the assembly of various nanoscale objects, such as metallic nanoparticles [29-33], carbon nanotubes [34], quantum dots or organic dyes [35], into highly-ordered structures.
Among plasmonic metamaterials, the use of anisotropic nanoparticles as building blocks, such as gold nanorods (AuNRs) [36-41], are especially intriguing as they have unique optical and electronic features, including wide range of optical extinction and strong plasmonic fields, as well as specific absorption owing to its anisotropic shape. Furthermore, the accurate assembly of AuNRs may lead to novel optoelectronic features that have never been observed in an individual AuNR as collective properties would emerge owing to the interparticle coupling upon forming the highly-ordered metamaterials. Importantly, these emerging collective properties can be rationally tuned by multiple geometric parameters, such as size, distance, and orientation [42, 43]. Along this direction, substantial efforts have been devoted to using DNA origami as template to direct the AuNR assembly and construct a variety of functional architectures, such as rectangular origamitemplated discrete AuNR dimer and helical superstructures with tailored chiroptical responses [44, 45]. More recently, Liu and coworkers used DNA origami template to create a range of reconfigurable 3D plasmonic nanosystems, which can respond to matter or external stimuli, including DNA strands [46], light [47], pH changes [48], and be monitored by CD spectrometry. Nevertheless, high-fidelity and high-yield assembly of AuNRs into elaborate nanoarchitectures still remains challenging, especially when the number of involved AuNRs increases. In this work, we designed and engineered three types of anisotropic AuNR clusters with different 3D spatial configurations using a rod-like DNA origami as the template (Fig. 1). By tuning the positions of capture strands on the surface of DNA origami nanorod (DOR), two-, three-, and four-AuNRs could be precisely arranged on the longitudinal direction of DOR to form parallel AuNR clusters in a high controllable manner. Then their optical properties were further investigated after the assembly.
|
Download:
|
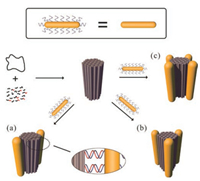
| Fig. 1. Schematic illustration of the rod-like DNA origami-directed assembly of AuNR cluster nanoarchitectures. A long single-stranded M13 DNA is folded by thermal annealing with a set of staple and capture strands to generate a rigid rodlike 54-helix bundle DNA origami template. The capture strands are extended from different sides of the DOR surface to achieve diverse AuNR assembly. On each side, multiple capture strands arranging evenly along the axis of DOR are used to robustly immobilize one AuNR. AuNRs modified with corresponding complementary DNA strands are assembled at the designated positions on the DNA origami via DNA hybridization, forming three types of parallel AuNR clusters: (a) dimers, (b) trimers and (c) tetramers, respectively. | |
In the material preparation, hexadecyltrimethyl ammonium bromide (CTAB), sodium borohydride (NaBH4), silver nitrate (AgNO3), tris(2-carboxyethyl)phosphine hydrochloride (TCEP) were supplied by Sigma. Gold (Ⅲ) chloride trihydrate (HAuCl4·3H2O) was supplied by J & K. L-Ascorbic acid was purchased from Macklin. Sodium dodecylsulfate (SDS) was purchased from Beyotime. Non-thiolated DNA sequences were bought from Sangon Biotech, while thiolated DNA sequences were obtained from DNA synthesizer.
In addition, DNA squences for folding the single-stranded M13mp18 DNA into origami rod were designed by using caDNAno software [49]. The AuNRs were prepared by following El-Sayed's method with some modifications [50]. Functionalization of the AuNRs with thiolated DNAs was conducted by using a previously reported salt aging method [51]. Meanwhile, the annealing processes of constructing DNA origami rod and the AuNR clusters followed Liedl's method [52] and Yan's method [43] respectively. After the assembly, DNA origami rods and the AuNR clusters were further purified by 1% agarose gel electrophoresis and recoverd by electroelution with dialysis bag. The details of DNA sequences, AuNR synthesis and functionalization, preparation and purification of DNA origami rod and AuNR clusters, sample characterizations including UV-vis spectroscropy, CD spectroscropy, agarose gel electrophoresis, AFM, and TEM imaging, etc. are presented in Supporting information.
Fig. 1 illustrated the design of DOR template and the assembly process of AuNR clusters. First, a rod-like 54-helix bundle DNA origami (45 nm × 20 nm) was synthesized following a typical method initiated by Rothemund [22], which can nicely match the length of gold nanorod used in this study. To further employ it as the template, two to four groups of the capture strands were introduced and extended from outside surface of the DNA origami rod. In detail, to achieve robust immobilization of AuNRs on DOR, multiple (five to ten, see Supporting information for the details) single-stranded overhangs for each group of capture strands were arranged along the longitudinal axis to form a binding site for immobilizing an AuNR (for details, see Supporting information). To construct AuNR clusters with different numbers, these capture strand groups were evenly arranged around the longitudinal axis of DORs with C2, C3, and C4 rotational symmetries. Then AuNRs (~45 nm × 12 nm on average) modified by thiolated DNAs with the sequences complementary to the single-stranded overhangs were synthesized following previously reported method [50, 51, 53]. Hence, DNA-modified AuNRs could be attached to the designated positions on the DNA origami rod through DNA hybridization. In this way, different types of anisotropic AuNR clusters can be successfully assembled, including AuNR dimers, trimers, and tetramers, in which all AuNRs are parallel to the axis of the DOR. From the top view of these parallel AuNR clusters, the AuNRs endpoints are located as a line segment, regular triangle, and rhombus, respectively.
With all designed DNA strands together, the rod-like DNA origami template was first assembled by folding the long M13mp18 scaffold strand with staple and capture strands at a ratio of 1:10:10 via a thermal annealing process from 80 ℃ to 25 ℃. Then the assembled product was characterized by agarose gel electrophoresis. As shown in Fig. 2a, a slightly faster mobility of the target DNA origami was observed in comparison with that of the pristine M13mp18 scaffold, which could be attributed to its tightly packed structure [54]. After removing the excess staple strands and unintended aggregates, the purified DNA origami was further visualized by transmission electron microscopy (TEM) and atomic force microscopy (AFM). As shown in Fig. 2, monodispersed rodlike nanostructures could be clearly observed (Figs. 2c and d, Figs. S2 and S3 in Supporting information), dimensions of which were highly in agreement with our design.

|
Download:
|
| Fig. 2. Characterizations of the DNA origami rods and the formation of AuNR clusters. (a) Agarose gel analysis of the self-assembled DOR stained by ethidium bromide and imaged under UV light. Lane 1: M13mp18 scaffold strand. Lane 2: The annealed products. The lowest bright band represents the excessive staple strands, followed by the DOR band above and some of unintended aggregates. (b) The formation of AuNR clusters by annealing the purified DOR template and the DNAmodified AuNRs together, which were determined by agarose gel electrophoresis. Lane 1: free DNA-modified AuNRs. Lane 2: the annealed AuNR cluster samples. The lowest band represents the excessive AuNRs, followed by the AuNR cluster band (dimer in this gel) above and some unintended aggregates. (c, d) AFM image (c) and TEM image (d) of the DNA origami rods after purification. | |
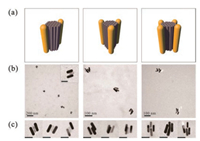
With the purified DOR in hand, DNA-modified AuNRs were further added in the solution and immobilized on the DOR template. To achieve high yield of the assembly, excess singlestranded DNA-functionalized AuNRs (ssDNA-AuNRs, with complementary sequences to the capture overhangs) were used and mixed with the DORs, followed by a multi-cycle annealing process over 25 h. After that, the assembled product of anisotropic AuNR clusters were analyzed by agarose gel electrophoresis (Fig. 2b and Fig. S4 in Supporting information). In all cases, sharp bands with lower gel mobility could be observed, which was located above the ssDNA-AuNR sample, indicating the good monodispersity of the assembled clusters. After purification of the target products from the gel by electroelution in a dialysis bag, the structures of anisotropic AuNR clusters were characterized by biological transmission electron microscopy (TEM) (Fig. 3 and Figs. S5-S10 in Supporting information). Even though the drying process on TEM grids caused deformation of the assembled plasmonic nanostructures in some degree, their native configurations were basically preserved and could be discriminated clearly, which mainly owes to the good rigidity of the DNA origami template. It is worth noting that, in the zoom-in TEM images, all those three types of AuNR architectures display corresponding side-by-side spatial configurations, which match very well with our design of parallel AuNR clusters. All these results demonstrate the excellent template role of rod-like DNA origami in precise control of the AuNRs' arrangement. Furthermore, the UV-vis spectral analysis of the AuNR clusters described above was carried out. Compared to the spectrum of ssDNA-AuNRs, absorptions of the organized plasmonic clusters was slightly blue-shifted, which may be aroused by interparticle coupling (Fig. S11 in Supporting information).
|
Download:
|
| Fig. 3. TEM characterization of the parallel AuNR clusters. (a) Schematic representation of each cluster construct. (b) Representative zoom-out TEM images of each cluster construct. The scale bar in the inset image is 50 nm. (c) Representative zoom-in TEM images revealing the precise arrangement of the AuNRs on the DNA origami template. Scale bars: 50 nm. | |
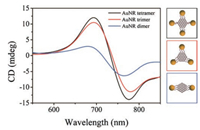
Finally, we implemented CD measurements using a circular dichroism spectrometer. Interestingly, chiral optical response of the assembled AuNR clusters was observed by the analysis of circular dichroism spectroscopy although they were designed to be highly symmetric architectures. Fig. 4 shows the CD spectra of these three types of parallel AuNR clusters. It is apparent that the anisotropic plasmonic clusters exhibit dramatical chiral optical responses with characteristic bisignate signatures, which can be attributed to the dipole-plasmon Coulomb interactions between AuNRs and chiral helices of double-stranded DNAs (dsDNA) [55, 56]. The left-handedness is induced by the parallel orientation of AuNRs relative to dsDNA in the template. While keeping the same concentration of AuNR clusters, it is found that the tetramers displayed the strongest CD signal, followed by the AuNR trimers. In contrast, AuNR dimers exhibit much weaker CD signal, which reflects the less extent of cooperative dipole-plasmon interactions between AuNR clusters and dsDNA helices [56].
|
Download:
|
| Fig. 4. CD spectra of the parallel AuNR clusters: dimers, trimers, and tetramers. The increasing trend of the CD intensity demonstrates its dependence on rod number. The concentrations of AuNR clusters are the same for these three samples during the measurements. | |
In summary, we have employed a DNA origami rod as the template to guide the construction of diverse anisotropic AuNR clusters with side-by-side 3D configurations. The remarkable programmability and addressability of origami structure ensures the precise spatial control of the plasmonic components, resulting in the well-defined AuNR dimer, trimer, and tetramer. With these newly engineered nanostructures, predictable and dramatical CD response at their optical frequencies was observed due to the dipole-plasmon interactions between AuNRs and dsDNA helices and the intensity of CD signal was found to be highly related to the number of AuNR in the clusters. Our work may broaden horizon in bottom-up fabrication of plasmonic nanoarchitectures with more complex 3D configurations and exploration of their emerging properties. Furthermore, it is also possible to realize precise 3D assemblies using other building blocks (e.g., quantum dots and magnetic nanoparticles) to construct more functional metamaterials and devices, which may perform as good platforms for biosensing, surface enhanced Raman scattering, drug delivery, and so on.
AcknowledgmentsThis work was supported by the National Natural Science Foundation of China (Nos. 21504053, 21661162001, 21673139, 91527304, 21722502), Shanghai Pujiang Talent Project (No. 16PJ1402700), the Innovation Fund from Joint Research Center for Precision Medicine set up by Shanghai Jiao Tong University & Affiliated Sixth People's Hospital South Campus (No. IFPM 2016B001), and the special program for collaborative innovation in Shanghai University of Medicine & Health Sciences (SPCI-17-15-001).
Appendix A. Supplementary dataSupplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.cclet.2018.04.020.
| [1] |
W. Li, Z.J. Coppens, L.V. Besteiro, et al., Nat. Commun. 6 (2015) 8379-8385. DOI:10.1038/ncomms9379 |
| [2] |
M. Ren, E. Plum, J.J. Xu, N.I. Zheludev, Nat. Commun. 3 (2012) 833-838. DOI:10.1038/ncomms1805 |
| [3] |
B. Luk'yanchuk, N.I. Zheludev, S.A. Maier, et al., Nat. Mater. 9 (2010) 707-715. DOI:10.1038/nmat2810 |
| [4] |
M.E. Stewart, C.R. Anderton, L.B. Thompson, et al., Chem. Rev. 108 (2008) 494-521. DOI:10.1021/cr068126n |
| [5] |
V.K. Valev, J.J. Baumberg, C. Sibilia, T. Verbiest, Adv. Mater. 25 (2013) 2517-2534. DOI:10.1002/adma.201205178 |
| [6] |
L. Gunnarsson, T. Rindzevicius, J. Prikulis, et al., J. Phys. Chem. B 109 (2005) 1079-1087. DOI:10.1021/jp049084e |
| [7] |
K.H. Su, Q.H. Wei, X. Zhang, et al., Nano Lett. 3 (2003) 1087-1090. DOI:10.1021/nl034197f |
| [8] |
W. Rechberger, A. Hohenau, A. Leitner, et al., Opt. Commun. 220 (2003) 137-141. DOI:10.1016/S0030-4018(03)01357-9 |
| [9] |
J. Grand, P.M. Adam, A.S. Grimault, et al., Plasmonics 1 (2006) 135-140. DOI:10.1007/s11468-006-9014-7 |
| [10] |
J. Prikulis, P. Hanarp, L. Olofsson, D. Sutherland, M. Käll, Nano Lett. 4 (2004) 1003-1007. DOI:10.1021/nl0497171 |
| [11] |
P. Hanarp, D. Sutherland, J. Gold, B. Kasemo, Colloids Surf. A 214 (2003) 23-36. DOI:10.1016/S0927-7757(02)00367-9 |
| [12] |
P. Hanarp, D. Sutherland, M. Kall, W. Bryant Garnett, Phys. Rev. Lett. 90 (2003) 057401. DOI:10.1103/PhysRevLett.90.057401 |
| [13] |
A.J. Haes, J. Zhao, S.L. Zou, et al., J. Phys. Chem. B 109 (2005) 11158-11162. DOI:10.1021/jp051178g |
| [14] |
E.M. Hicks, O. Lyandres, W.P. Hall, et al., J. Phys. Chem. C 111 (2007) 4116-4124. |
| [15] |
P.F. Zhan, P.K. Dutta, P.F. Wang, et al., ACS Nano 11 (2017) 1172-1179. DOI:10.1021/acsnano.6b06861 |
| [16] |
N.C. Seeman, Nature 421 (2003) 427-431. DOI:10.1038/nature01406 |
| [17] |
Y. Krishnan, F.C. Simmel, Angew. Chem. Int. Ed. 50 (2011) 3124-3156. DOI:10.1002/anie.200907223 |
| [18] |
X.R. Wu, C.W. Wu, F. Ding, et al., Chin. Chem. Lett. 28 (2017) 851-856. DOI:10.1016/j.cclet.2017.01.012 |
| [19] |
X.R. Wu, C.W. Wu, C. Zhang, Chin. J. Polym. Sci. 35 (2017) 1-24. DOI:10.1007/s10118-017-1871-3 |
| [20] |
X. Li, L. Hong, T. Song, et al., J. Biomed. Nanotechnol. 13 (2017) 747-757. DOI:10.1166/jbn.2017.2383 |
| [21] |
H.Q. Wang, Z.X. Deng, Chin. Chem. Lett. 26 (2015) 1435-1438. DOI:10.1016/j.cclet.2015.10.019 |
| [22] |
P.W.K. Rothemund, Nature 440 (2006) 297-302. DOI:10.1038/nature04586 |
| [23] |
F. Hong, F. Zhang, Y. Liu, H. Yan, Chem. Rev. 117 (2017) 12584-12640. DOI:10.1021/acs.chemrev.6b00825 |
| [24] |
E.S. Andersen, M.D. Dong, M.M. Nielsen, et al., Nature 459 (2009) 73-77. DOI:10.1038/nature07971 |
| [25] |
S.M. Douglas, H. Dietz, T. Liedl, et al., Nature 459 (2009) 414-418. DOI:10.1038/nature08016 |
| [26] |
D.R. Han, S. Pal, J. Nangreave, et al., Science 332 (2011) 342-346. DOI:10.1126/science.1202998 |
| [27] |
E. Benson, A. Mohammed, J. Gardell, et al., Nature 523 (2015) 441-444. DOI:10.1038/nature14586 |
| [28] |
G. Tikhomirov, P. Petersen, L. Qian, Nature 552 (2017) 67-71. DOI:10.1038/nature24655 |
| [29] |
B.Q. Ding, Z.T. Deng, H. Yan, et al., J. Am. Chem. Soc. 132 (2010) 3248-3249. DOI:10.1021/ja9101198 |
| [30] |
S. Pal, Z.T. Deng, B.Q. Ding, H. Yan, Y. Liu, Angew. Chem. Int. Ed. 49 (2010) 2700-2704. DOI:10.1002/anie.201000330 |
| [31] |
W.Y. Liu, M. Tagawa, H.L. Xin, et al., Science 351 (2016) 582-586. DOI:10.1126/science.aad2080 |
| [32] |
M.J. Urban, P.K. Dutta, P.F. Wang, et al., J. Am. Chem. Soc. 138 (2016) 5495-5498. DOI:10.1021/jacs.6b00958 |
| [33] |
Y. Tian, Y.G. Zhang, T. Wang, et al., Nat. Mater. 15 (2016) 654-661. DOI:10.1038/nmat4571 |
| [34] |
H.T. Maune, S.P. Han, R.D. Barish, et al., Nat. Nanotechnol. 5 (2010) 61-66. DOI:10.1038/nnano.2009.311 |
| [35] |
R. Schreiber, J. Do, E.M. Roller, et al., Nat. Nanotechnol. 9 (2014) 74-78. DOI:10.1038/nnano.2013.253 |
| [36] |
H.D. Cui, D.H. Hu, J.N. Zhang, et al., Chin. Chem. Lett. 28 (2017) 1391-1398. DOI:10.1016/j.cclet.2016.12.038 |
| [37] |
A. Pitchaimani, N.T.D. Thanh, L. Maurmann, et al., J. Biomed. Nanotechnol. 13 (2017) 417-426. DOI:10.1166/jbn.2017.2362 |
| [38] |
M. Singh, D.C.C. Harris-Birtill, Y. Zhou, et al., J. Biomed. Nanotechnol. 12 (2016) 481-490. DOI:10.1166/jbn.2016.2196 |
| [39] |
V.S. Marangoni, J. Cancino-Bernardi, V. Zucolotto, J. Biomed. Nanotechnol. 12 (2016) 1136-1158. DOI:10.1166/jbn.2016.2218 |
| [40] |
N. Zhang, D.X. Zhu, F. Li, et al., J. Biomed. Nanotechnol. 13 (2017) 134-143. DOI:10.1166/jbn.2017.2330 |
| [41] |
J.H. Zhou, Y.Y. Wang, L. Zhang, X.M. Li, Chin. Chem. Lett. 29 (2018) 54-60. DOI:10.1016/j.cclet.2017.09.003 |
| [42] |
X.H. Huang, S. Neretina, M.A. El-Sayed, Adv. Mater. 21 (2009) 4880-4910. DOI:10.1002/adma.200802789 |
| [43] |
S. Pal, Z.T. Deng, H.N. Wang, et al., J. Am. Chem. Soc. 133 (2011) 17606-17609. DOI:10.1021/ja207898r |
| [44] |
X. Lan, Z. Chen, G.L. Dai, et al., J. Am. Chem. Soc. 135 (2013) 11441-11444. DOI:10.1021/ja404354c |
| [45] |
X. Lan, X.X. Lu, C.Q. Shen, et al., J. Am. Chem. Soc. 137 (2015) 457-462. DOI:10.1021/ja511333q |
| [46] |
A. Kuzyk, R. Schreiber, H. Zhang, et al., Nat. Mater. 13 (2014) 862-866. DOI:10.1038/nmat4031 |
| [47] |
A. Kuzyk, Y.Y. Yang, X.Y. Duan, et al., Nat. Commun. 7 (2016) 10591. DOI:10.1038/ncomms10591 |
| [48] |
A. Kuzyk, M.J. Urban, A. Idili, F. Ricci, N. Liu, Sci. Adv. 3 (2017) e1602803. DOI:10.1126/sciadv.1602803 |
| [49] |
S.M. Douglas, A.H. Marblestone, S. Teerapittayanon, et al., Nucleic Acids Res. 37 (2009) 5001-5006. DOI:10.1093/nar/gkp436 |
| [50] |
B. Nikoobakht, M.A. El-Sayed, Chem. Mater. 15 (2003) 1957-1962. DOI:10.1021/cm020732l |
| [51] |
M.R. Jones, R.J. Macfarlane, B. Lee, et al., Nat. Mater. 9 (2010) 913-917. DOI:10.1038/nmat2870 |
| [52] |
A. Kuzyk, R. Schreiber, Z.Y. Fan, et al., Nature 483 (2012) 311-314. DOI:10.1038/nature10889 |
| [53] |
X.M. Qu, D. Zhu, G.B. Yao, et al., Angew. Chem. Int. Ed. 56 (2017) 1855-1858. DOI:10.1002/anie.201611777 |
| [54] |
X.B. Shen, C. Song, J.Y. Wang, et al., J. Am. Chem. Soc. 134 (2012) 146-149. DOI:10.1021/ja209861x |
| [55] |
C.C. Rao, Z.G. Wang, N. Li, et al., Nanoscale 7 (2015) 9147-9152. DOI:10.1039/C5NR01634G |
| [56] |
X. Lan, Q.B. Wang, Adv. Mater. 28 (2016) 10499-10507. DOI:10.1002/adma.v28.47 |
 2019, Vol. 30
2019, Vol. 30 

