b Hangzhou Pharmaceutical Group Co., Ltd., Hangzhou 311100, China
NL-101 (Fig. 1), as one of the new model nitrogen mustard (NM) drugs, has been already utilized as an alkylating agent for treatment of the lymphatic and myeloid system cancer. Biological activity of NM aims to cause linkages between strands of DNA to inhibit synthesizing DNA and RNA [1]. It has been proposed that the drug-combined adducts are involved with the distal guanine bases in the strands of 50-GNC sequences [2]. Besides, the pivotal NM cytotoxicity within GNC sequences is DNA-DNA inter-strand cross-linking between the N7 positions of distal guanine nucleobases and drugs [3-5]. Since the efficacy and toxicity of anticancer drugs are related to parent drug and/or metabolite concentrations in body fluids and tissues, liquid chromatography (LC) has been employed for analyzing such complicate samples.
|
Download:
|
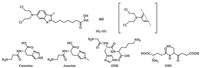
| Fig. 1. Structures of NL-101 and peptides. | |
In recent years, electrospray ionization mass spectrometry (ESIMS) has become as one of the focal points in scientific research. Due to its high vacuum environment, ESI-MS could measure compounds by isolation and storing them with no complicating interferences by solvent effects, which provides a profound understanding of the intrinsic mechanism in organic reaction [6-9]. In addition, the subsequent tandem mass spectrometry (MS/ MS) reactions under collision-induced dissociation (CID) could provide further details about the structure of target compounds, which are protonated at the ionization process at the initial stage of a typical ESI-MS experiment. As a result, high performance liquid chromatography (HPLC) combined with MS has a pervasive application in analysis of complicated samples.
It is noteworthy that NM drugs hold several potential risks for the patients, owing to their low selectivity and carcinogenicity [10-12]. The accumulation of mutations within DNA strand and protein-drug adducts would increase the probability of malfunctions in the metabolism of the cell, usually resulting in apoptosis [12-14]. However, previous studies [10-14] demonstrated ambiguous structural information about the biomolecule and the drug adducts.
In this work, high performance liquid chromatography electrospray tandem mass spectrometry (HPLC/ESI-MS/MS) was utilized to investigate covalent binding sites of NL-101 with amino acids or polypeptides. The binding sites of amino acids-NL and peptides-NL were analyzed by CID technique in positive electrospray ionization mode (ESI+). The experimental section was shown in Supporting information. The NL-101 was characterized by ESI-MS as shown in Figs. S1–S3 (Supporting information). The mixture of NL-101 and amino acids and peptides were incubated at 37 ℃ for 4 h to obtain enough abundance to identify.
Due to its nonpolar side chain with only two functional groups available for addition, valine was chosen as a model. Incubation of NL-101 with valine at pH 7.3 revealed two adducts at both sites, the amino group combined with NL-101 adducts (Val-NH-NL), the carboxyl group combined with NL-101 adduct (Val-COO-NL), eluting at 14.44 min and 15.36 min respectively (Fig. 2A). The characteristic chlorine isotope pattern of the adducts was authenticated as Val-NL(Cl) at m/z 496. The fragment ion m/z 478 in both CID spectra was considered as Val-NL(OH) which was produced by the replacement of chloride with hydroxyl group. CID was utilized to distinguish the location of the binding sites and demonstrated the different fragmentation patterns. The former spectrum showed two losses of 18 Da and they were observed at m/z 442, 460 and 478 and data of high-resolution were shown in Table S1 (Supporting information). Fragment ion at m/z 442 might represent as piperazine ring, a unique fragment ion formed from the amino group adducts (Fig. 2B).

|
Download:
|
| Fig. 2. (A) IC of Val-NL (parent ions at m/z 496) adducts formed at pH 5.3, 7.3 and 9.3. (B) CID product ion spectra of Val-NL(Cl) adducts. | |
When the incubation experiment was performed at pH 5.3, there was only one adduct product observed and considered as Val-COO-NL. According to the HPLC results (Fig. 2A), there were two adduct products, one dominant and one minor. This phenomenon may be attributed to activation of the carboxyl and amino groups at different pH conditions. The amino group is inclined to obtain H+ and became electrophilic site, which is difficult to combine with another electrophilic aziridinium ion. When the surrounding is basic (pH 9.3), the amino group could be more active comparing with the pH 5.3 and 7.3. On the other hand, the carboxyl group could lose one H+ altering as nucleophilic site, and is more available for adduct formation. According to the analysis above, the varied binding sites of amino acid-NL (Cl) appearing at different pH conditions could be interpreted.
Meanwhile, valine methyl ester, Val(ester), was treated to support the results above since only the amino group left actively. Figs. S4 and S5 (Supporting information) illustrated a single peak eluted at 15.08 min in the ion chromatograms (IC) had the similar fragmentation pattern as that of Val-NH-NL. The fragmentation patterns between the amino and carboxyl group adducts had also been verified upon alanine, leucine, phenylalanine and its ester compound (Figs. S6–S12 in Supporting information).
Sulfhydryl group is one of the strongest nucleophiles in amino acids, which is highly reactive group in several proteins, enzymes. Meanwhile, disulfide bond plays a valuable role by crosslinking proteins and increases the rigidity of proteins and functions to confer proteolytic resistance [15, 16]. Therefore, analysis of Cys-NL (Cl) and Met-NL(Cl) adducts could supply a common tool for further study upon peptides or proteins. Both Cys and Met appeared high selectivity to covalently bind to NL-101.
Adducts of Met-NL(Cl) separated two adducts at the experimental conditions (Figs. S13 and S14 in Supporting information). Besides, the MS/MS spectra of Met-NL(Cl) behaved utterly different, corresponding to their binding sites (Fig. 3). The first adduct eluting at 5.34 min has a unique fragment ion, which was considered as Met-S-NL. The most abundant fragment ion, m/z 427, was considered as C20H32ClN4O2S+, which can be classified as the protonated thioether on NL-101. And CID spectrum of the second adduct is similar to fragmentation pattern of the Val-NH-NL and naturally categorized as Met-NH-NL.

|
Download:
|
| Fig. 3. CID spectra of Met-NL (parent ions at m/z 528 Da) adducts. | |
Incubated NL-101 with Cys at experimental conditions, adducts Cys-NL(Cl) formed at acidic condition (pH 5.3) eluted at 13.37 min and 15.21 min (Figs. S15 and S16 in Supporting information). With the incubation condition getting alkaline, only one adduct left and its CID displayed particular fragment ions, m/z 413, 421 and 439 (Fig. S15B in Supporting information). These can be illustrated by the competition between the sulfhydryl group and the amino group. Therefore, these two adducts obtained under pH 5.3 condition could be counted as Cys-S-NL and Cys-NH-NL. Cys-NHNL adduct, which was alkylated at the amino group, showed a series losses of 18 Da product ions at m/z 446, 464 and 482. These fragment ions corresponded to the formation of aziridinium ion which was attributed to the nucleophilic attack of the amino group on the ethylene moiety of NL-101. Fragment ion at 464 Da might be considered as piperazine ring, a unique fragment ion formed from the amino group adducts. The CID spectra of Cys-S-NL showed unique fragment ions, which implied the binding of NL-101 to the sulfhydryl group.
Histidine has an imidazole ring as a side chain, whose pKa is 6.04, and is naturally deprotonated at physiological condition (pH 7.3). Three His-NL(Cl) adducts at m/z 534 eluted at 3.81, 4.66 and 5.15 min at the experimental conditions (Fig. S17A in Supporting information), which could stand for adducts on the site of the carboxyl, amino and the nitrogen atoms of the imidazole ring. According to the CID experiments, each peak revealed their characteristic fragment ions, which was useful for locating the binding sites. The adduct eluted at 3.81 min was identified as an amino group adduct, His-NH-NL, by the fragment ion at m/z 480 Da, in analogy to the piperazine ring product ion found for the amino alkylated valine, Val-NH-NL (Fig. 2B). The second peak represented the carboxyl group adduct, His-COO-NL, by the similar fragment ions coordinating with Val-COO-NL (Fig. 2B). Therefore, the last eluted peak with several unique fragment ions stand for the imidazole nitrogen adduct, His-im-NL. Particularly, the product ion at m/z 427 was composed of imidazole ring and NL-101 (Fig. S17B in Supporting information). However, there was not enough information, based on these fragment patterns, to locate binding site on the specific amino groups in imidazole ring.
Glutamic acid (Glu) has an acidic side chain with a pKa of 4.15. At experimental conditions, adducts of Glu-NL(Cl) were observed and eluted at 12.36 and 16.25 min, which were considered as Glu-COONL and Glu-NH-NL, respectively (Fig. S18 in Supporting information). The CID spectrum of Glu-COO-NL illustrated high intensity of fragment ions at m/z 490 and m/z 508, which can explain by the formation of a six-membered ring by losing a H2O of glutamic acid on the NL-101 (Fig. S19 in Supporting information). However, only based on the CID spectra, the authentic binding site of Glu-COO-NL adduct cannot be determined. On the other hand, the spectrum of Glu-NH-NL exhibited the particular fragment ions, m/z 446 and m/z 464. The loss of a H2O from Glu-NH-NL adduct suggests the ring formation upon fragmentation (Fig. S20 in Supporting information). The double carboxyl groups that led to the ring formation might be the reason for the slightly different fragmentation patterns comparing with Val-NL(Cl).
Incubation of NL-101 with GSH at experimental conditions gave the same information of IC about the two GSH-NL(Cl) adducts eluting at 7.73 min and 9.68 min (Fig. S21 in Supporting information). These GSH-NL(Cl) adducts were verified as products on the amino and sulfhydryl groups. Based on CID spectra, identification of the binding sites could be supported by fragment ions. The first adduct eluting at 7.73 min produced more fragment ions and the fragmentation pattern of m/z at 650, 668 and 686 is in analogy to the piperazine ring product ion found for Val-NH-NL, among which fragment ion at m/z 650 could be exploited to prove the binding site at the amino site. The most abundant fragment ion at m/z 508 could be explained by the six-membered ring formation, and fragment ion at m/z 490 could be treated as replacement of chloride with hydroxyl group (Fig. S22 in Supporting information). The second adduct appeared to be the sulfhydryl group adduct, whose CID spectrum showed unique product ions that allowed a rapid and persuasive assignment (Fig. 4).

|
Download:
|
| Fig. 4. CID product ion spectra of GSH-NL (parent ion at m/z 686) adducts. | |
Carnosin (β-alanyl-L-histidine) and anserine (β-alanyl-N-methylhistidine) are both highly concentrated in muscle and brain tissues [17], which are able to scavenge reactive oxygen species, and act as anti-glycating agent, reducing the rate of formation of advanced glycation end-products (substances that can be a factor in the development or worsening of many degenerative diseases, such as diabetes, atherosclerosis, chronic renal failure, and Alzheimer's disease) and so on [18, 19].
Two Car-NL(Cl) adducts were formed under incubation experiment but no adducts at pH 5.3. However, the two adducts were poorly separated, and even adjusting the LC gradient did not attain a good separation (Fig. S23 in Supporting information). The adduct ion eluted at 4.89 min was identified as adduct at the amino group of β-alanine because MS/MS experiment represented a unique fragment ion at m/z 551, which is a pivotal ion for recognition of binding on amino group, which also appeared on the other-NH-NL adducts. The second adduct ion eluted at 5.04 min was considered as adduct on the imidazole-ring, Car-im-NL, due to the unique fragment ions like m/z 453 and m/z 427 (Fig. S24 in Supporting information). The high intensity of product ion at m/z 588 could be explained by the intramolecular cyclization reaction resulting in the formation of a seven-membered ring and the replacement of chloride with hydroxyl group as the product ion at m/z 570 (Fig. S25 in Supporting information).
Tripeptide GHK-Cu can promote activation of wound healing, attraction of immune cells, stimulation of collagen and glycosaminoglycan synthesis in skin fibroblasts and promotion of blood vessels growth [20]. Meanwhile, it has been revealed that GHK-Cu is able to modulate expression of a large number of human genes, generally reverse gene expression to a healthier state [21].
Tripeptide GHK-Cu can promote activation of wound healing, attraction of immune cells, stimulation of collagen and glycosaminoglycan synthesis in skin fibroblasts and promotion of blood vessels growth [20]. Meanwhile, it has been revealed that GHK-Cu is able to modulate expression of a large number of human genes, generally reverse gene expression to a healthier state [21].
GHK, was incubated with NL-101 resulting in three GHK-NL(Cl) adducts at experimental conditions, which suggests its independence of experimental pH. GHK-NL(Cl) adducts eluted at 5.23, 5.58 and 6.48 min at pH 7.3 were identified as binding to the different site of amino groups (Fig. S26A in Supporting information). Each spectrum had characteristic ions helping verify the binding sites. The particular ion on the first CID spectrum of GHK-NL(Cl) was at m/z 657, and that of the second CID was at m/z 555, 648 and 666 (Fig. S26B in Supporting information). However, only based on the CID spectra, the authentic binding site of GHK-NH-NL adducts cannot be determined. Possible reason of the differences between the two CID spectra could be the diversity of proton affinity of binding nitrogen site. Naturally, imidazole ring is more nucleophilic than other amino group in GHK. Thus, the highest intensity of three peaks was speculated as adduct at the imidazole ring, GHK-im-NL. The binding sites, such as amino, carboxyl and R group, were confirmed by tandem mass spectrometry experiments. Amino acid-NH-NL often produce more fragment ions; amino acid-COO-NL display fewer fragment ions. And when the binding site was located at R group, its CID spectra showed distinguish fragmentation pattern and could have a rapid identification. HPLC/MS/MS has been documented as an effective method for the characterization of nitrogen mustard drug-binding small peptides. Through this specific analysis, the results could provide a transparent view of structural information about the NL- 101 adducts and present possible explanations of the cytotoxic and side effects of NL-101 after administered by intravenous injection.
In conclusion, HPLC/ESI-MS/MS was exploited to analyze the adducts of amino acids and peptides with NL-101. The functional group of NL-101 suggested that the reaction between amino acids and NL-101 involved several binding sites, such as amino, carboxyl and R group, which were confirmed by CID. Amino acid-NH-NL often produces more fragment ions; amino acid-COO-NL displays fewer fragment ions. And when the binding site was located at R group, its CID spectra showed distinguish fragmentation pattern and could have a rapid identification. Through this specific analysis, the results could provide a transparent view of structural information about the NL-101 adducts and present possible explanations of the cytotoxic and side effects of NL-101 after administered by intravenous injection.
AcknowledgmentThis work was supported by the National Natural Science Foundation of China (Nos. 21327010, 21372199).
Appendix A. Supplementary dataSupplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.cclet.2017.12.023.
| [1] |
R. Bansal, P.C. Acharya, Chem. Rev. 114 (2014) 6986-7005. DOI:10.1021/cr4002935 |
| [2] |
S. Balcome, S. Park, D.Q. Dorr, et al., Chem. Res. Toxicol. 17 (2004) 950-962. DOI:10.1021/tx0499463 |
| [3] |
S.M. Rink, P.B. Hopkins, Biochemistry 34 (1995) 1439-1445. DOI:10.1021/bi00004a039 |
| [4] |
J.T. Millard, R.J. Spencer, P.B. Hopkins, Biochemistry 37 (1998) 5211-5219. DOI:10.1021/bi972862r |
| [5] |
R.F. Struck, R.L. Davis, M.D. Berardini, et al., Cancer Chemoth. Pharm. 45 (2000) 59-62. DOI:10.1007/PL00006744 |
| [6] |
R. Jirásko, M. Holcapek, Mass. Spectrom. Rev. 30 (2011) 10139-1036. |
| [7] |
C. Iacobucci, S. Reale, F.D. Angelis, Angew. Chem. Int. Ed. 55 (2016) 2980-2993. DOI:10.1002/anie.201507088 |
| [8] |
R.J. O'Hair, N.J. Rijs, Acc. Chem. Res. 48 (2015) 329-340. DOI:10.1021/ar500377u |
| [9] |
D. Schröder, Acc. Chem. Res. 45 (2012) 1521-1532. DOI:10.1021/ar3000426 |
| [10] |
L.J. Kinlen, Am. J. Med. 78 (1985) 44-49. |
| [11] |
S. Balcome, S. Park, D.Q. Dorr, et al., Chem. Res. Toxicol. 17 (2004) 950-962. DOI:10.1021/tx0499463 |
| [12] |
R. Loeber, E. Michaelson, Q.M. Fang, et al., Chem. Res. Toxicol. 21 (2008) 787-795. DOI:10.1021/tx7004508 |
| [13] |
D. Noort, A.G. Hulst, R. Jansen, Arch. Toxicol. 76 (2002) 83-88. DOI:10.1007/s00204-001-0318-2 |
| [14] |
A. Groehler, P.W. Villalta, C. Campbell, et al., Chem. Res. Toxicol. 29 (2016) 190-202. DOI:10.1021/acs.chemrestox.5b00430 |
| [15] |
C.S. Sevier, C.A. Kaiser, Nat. Rev. 3 (2002) 836-847. |
| [16] |
M. Ruoppolo, F. Vinci, T.A. Klink, et al., Biochemistry 39 (2000) 12033-12042. DOI:10.1021/bi001044n |
| [17] |
J.A. Zapp, D.W. Wilson, J. Biol. Chem. 126 (1938) 19-27. |
| [18] |
O.I. Aruoma, M.J. Laughton, B. Halliwell, Biochem. J. 264 (1989) 863-869. DOI:10.1042/bj2640863 |
| [19] |
G.I. Klebanov, Y. Teselkin, I.V. Babenkova, et al., Membr. Cell Biol. 12 (1998) 89-99. |
| [20] |
L. Pilgeram, Cardiovasc. Eng. 10 (2010) 78-83. DOI:10.1007/s10558-010-9092-1 |
| [21] |
L. Pickart, J. Biomater, Sci. Polymer Ed. 19 (2008) 969-988. |
 2019, Vol. 30
2019, Vol. 30 

