b College of Life Science, Key Laboratory of Medicinal Chemistry and Molecular Diagnosis of the Ministry of Education, Hebei University, Baoding 071002, China;
c University of Chinese Academy of Sciences, Beijing 100049, China
The design, synthesis and application of small molecules that can selectively bind at specific nucleic acid sequences and structures in RNA have attracted significant interest within the chemistry and biology community [1-11]. They could potentially be used as sequence/structure-specific probes or pharmaceuticals that are capable of disturbing RNA functions for understanding or controlling genetic information expression. To this end, decades of researches have introduced many types of RNA ligands/binders that can specifically target RNA of interest (ROI) with diverse activity ranging from 1 nmol/L to 1 mmol/L [12-24]. However, despite the great potential and numerous effort in this field, finding selective ligands for a specific sequence and with drug-like properties still remains a challenge. Many known RNA-binding molecules are polycationic, thus binding tightly with polyanionic RNA but with limit selectivity. Therefore, developing organic and/or organometallic molecules that can specifically bind RNA with high affinity is a challenge that must be addressed for RNA to be considered a feasible drug target. Organic ligands bound with metal centers are highly integrated systems that can be functionalized to control a variety of bimolecular processes [25-29]. Unlike traditional aminoglycosides, polypeptides, polycyclic aromatic molecules and other nucleic acid binders with a significant amount of cationic charge or aromatic density, the metal ions bind to nucleobases and/or phosphates via specific coordination bonds. Those metal complexes can be employed as potential therapy and diagnosis. However, contrary to the extensive investigation of organometallics that bind with DNA, their RNA binding proprieties remained largely unexplored [30-34].
Previously, we have explored several functionalized deoxygenated polypeptide/peptoids (DOPPs) that can specifically bind to folded RNA structures, such as (UUU)n repeats [35-36]. Polyamines with chiral side histidine chains could sterically shied the cationic charges in RNA backbones while remaining its capacity to engage in catalytic cleavage of phosphate linkages [37-39]. Furthermore, we have examined whether polyamine-metal complexes with zinc, copper and iron can be utilized in this process [36]. The preliminary results indicated that organozinc/copper/iron complexes could bind with RNA with high affinity and specificity. Based on this, we have initiated a new project to investigate whether the organic ligand that bound with metal centers can be optimized to direct binding specificity. To examine these ideas, two types of dinuclear metal complexes based on 2, 7-disubstituted 1, 8-naphthalenediol ligands have been prepared [40], and their binding to the folded RNAs associated with HIV, namely TAR (Trans-Activation Response) and RRE (Rev Response Element), has been studied (Fig. 1). TAR and RRE represent two of the most significant secondary structures in RNAs [41-45]. Two noteworthy features, a stem and loop with bulges that are sites for protein binding, make them promising targets to inhibit the viral replication and for developing antiviral compounds [43]. The results presented here demonstrated that functionalized dinuclear metal complexes can be readily synthesized, and that the binding mode, i.e. the affinity and specificity with TAR and RRE, are sensitive to alteration of the organic ligands and metals. We also performed a computational study aimed at better identifying the bind of these small organometallic complexes to TAR RNA.

|
Download:
|
| Fig. 1. Dinuclear metal complex scaffolds (A), secondary structures of TAR (B) and RRE RNA (C). | |
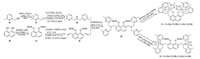
The synthesis of complexes 1 and 2 was carried out starting from commercial available compounds 3a and 3b (Scheme 1). Compound 3a was converted to dipyridinyl methyl amine 3d in 61% yield. The diol 3b was protected with chloromethyl methyl ether (MOMCl), which facilitated further α-lithiation and formylation to generate the dicarbaldehyde 3f in 40% yield. The reductive condensation of 3f with aforementioned 3d produced diamine 3g, from which two series of dipyridinyl-N bridged dinuclear metal complexes 1 and 2 were synthesized in moderate to good yields.
|
Download:
|
| Scheme 1. Synthesis of dinuclear metal complexes 1 and 2. | |
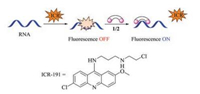
In order to investigate the interaction mode between the synthetic dinuclear metallic ligands 1 and 2 and RNA, fluorescence displacement experiments were employed [46]. The intrinsic fluorescence intensity of RNA is negligible. However, when a fluorescent probe, like ICR-191 that we used here, intercalated with RNA, the fluorescence intensity of the ligand will be quenched (fluorescence OFF). Upon the addition of external ligands, the binding sites of the RNA available for fluorescent probe will be decreased, and hence restore the signal (fluorescence ON). Therefore, this fluorescence displacement experiments can be used to probe the interaction of complexes with RNA (Fig. 2).
|
Download:
|
| Fig. 2. Fluorescence displacement experiments with ICR. | |
In our experiment, as illustrated in Fig. 3, the fluorescence intensities of ICR-191 bound to TAR RNA at 1 mm show remarkable decreasing trends with the increasing concentration of the dimetallic complexes 1a and 2b, respectively, indicating that some ICR molecules were released into solution after the exchange with the dimetallic complex, and resulted in the fluorescence recovering of ICR. Similar phenomenon was observed for RRE RNA and small molecules (vide infra).

|
Download:
|
| Fig. 3. Fluorescence spectra of TAR RNA-ICR upon the addition of complexes 1a (A) and 2b (B). The arrows show the change upon the increasing amount of complex concentration. Inset: plot of F0/Fvs. [complex] for the titration of the complex to ICR-191. Plot of lg(F–F0)/F0 vs. lg[[Dt] – n(F–F0)/F [Rt]] for the titration of complexes 1a (C) and 2b (D) to ICR. Binding assay conditions: The fluorescence displacement experiments were performed in Na2HPO4-NaH2PO4 buffer solution (20 mmol/L, pH 7.0 at 25 ℃, containing 20 mmol/L NaCl) with the concentrations of both ICR 191 and TAR RNA at 1 mmol/L. All experiments were performed on a fluorescence spectrophotometer Hitachi F7000. The excitation wavelength for ligand ICR 191 was set at 411 nm, emission wavelength was set at 420 nm, scaning range 420-600 nm. The slit width was set at (5, 5) nm, scanning speed 1200 nm/min and photomultiplier voltage was set as 600 V, response: 2.0 s. | |
These observations may be interpreted as the intercalation of the dimetallic complex with RNA. The recovering of ICR released from RNA by complexes 1a and 2b is in agreement with the linear Eq. (1):
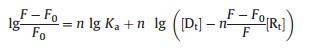
|
where [Rt] and [Dt] represent the overall concentrations of ICRRNA complex and the dinuclear metallic ligand. Thus the Ka and n values were obtained for the curve [46] and are listed as below (Table 1), from which a specific or general affinity for TAR RNA was observed with compounds 1a and 2b. For a comparison, we also tested 3-amino-6-methyl-N-(3-(trifluoromethyl)phenyl)thieno [2, 3-b]pyridine-2-carboxamide 3, a reported small molecule inhibitor for TAR RNA [47-48]. Again, compound 2b shows preferably binding affinities towards TAR RNA.
|
|
Table 1 Binding sites n and binding constants Ka for complexes 1a, 2b and 3. |
In order to obtain insight into the preferred binding location and help deepen the understanding of the dinuclear metallic ligands-RNA interaction, the molecular docking technique was used to dock complexes 1 and 2 into TAR RNA. AutoDock 4.2 using the Lamarckian Genetic Algorithm (LGA) for the prediction of binding affinity and searching for the optimum binding site together with the AutoDock Tools (ADT) were employed to set up and perform blind docking calculations of the eight dinuclear complexes [49]. The structure of RNA with sequence GCCAGAUUUGAGCCUGGGAGCUCUCUGGC (PDB ID: 1QD3, a sequence used in oligonucleotide study) obtained from the Protein Data Bank (www.rcsb.org/pdb) was constructed using AutoDock 4.2 package to study the RNA-binding properties. The copper parameters, a vdW radii of 0.96 Å and a vdW well depth of 0.01 kcal/mol, used in the docking calculation were taken from ref. [50]. The coordinates of complexes 1a and 2b were taken from their crystal structures as a CIF file and converted to the PDB format using Mercury software 3.10. The receptor (TAR RNA) and the ligands (dinuclear complexes) files were prepared using AutoDock Tools. The heteroatoms including water molecules were deleted and polar hydrogen atoms and Kollman charges were added to the receptor molecule. All other bonds were allowed to be rotatable. In the docking analysis, the binding site was assigned across all of the minor and major grooves of the RNA molecule, which was enclosed in a box with the number of grid points in x × y × z directions, 70 × 70 × 70 and a grid spacing of 0.375 Å. Initially, AutoGrid was run to generate the grid map of various atoms of the ligands and receptor. After the completion of the grid map, AutoDock was run by using autodock parameters as follows: GA population size, 150; maximum number of energy evaluations 2, 500, 000; and the number of generations 27, 000. A total of 10 runs were carried out. A maximum of 30 conformers were considered for each molecule, and the root-mean-square (RMS) cluster tolerance was set to 2.0 Å in each run. All calculations were performed on an Intel Core i5 based machine running Windows as the operating system. For each of the docking cases, the lowest energy docked conformation, according to the Autodock scoring function, was selected as the binding mode. Visualization of the docked pose was done by using PyMOL (The PyMOL Molecular Graphics System, Version 2.0, Schrödinger, LLC) molecular graphics program [51].
With the purpose of gaining more information concerning the favorably ligands of 2b vs. 1a towards TAR RNA, the binding sites and the binding poses of the two dimetallic complexes in the docking model with 1QD3 are shown in Fig. 4, from which it can be seen that the binding poses of the two dimetallic complexes are different. While complex 1a (O62) can interact with the NH7 (A20) (1.868 Å), 2b forms four different bindings: OP2 (U38)-N3, 2.878 Å; OP1 (G36)-O3, 2.678 Å, OP1 (G36)-O1, 2.936 Å; OP2 (G21)-O2, 2.894 Å, leading to an increase of the electrostatic energy and thus the stronger binding affinity of complex 2b to TAR RNA, which coincides with the results obtained from the experimental studies (Table 2). While the AutoDock calculation indicated 2b did not directly interact with TAR RNA, we predicted a hydrogen bond network might be involved via water molecule(s) [52-54]. It should be pointed out that the modeling studies of the interaction only allow a comparison between the two potential binding ligands. However, the above spectroscopy experiments together with the molecular docking studies can provide a comprehensive understanding of the RNA binding activities, proving that the dinuclear metallic complexes can strongly recover the intrinsic fluorescence of ICR through the static binding mechanism with the ratio of the dicopperⅡ complex being close to 1:1 while the dicopperⅡ being 1.5:1, and the most possible binding site is in the proximity of stem area. Thus, the reason that compounds 1a and 2b bind preferably with TAR may be ascribed to the longer distance between upper stem and lower stem in RRE.
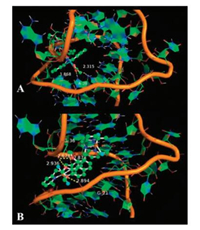
|
Download:
|
| Fig. 4. Visualization of the docked poses of complexes 1a (A) and 2b (B) with TAR RNA. The wireframe model of TAR with the dimetallic complexes (ball and stick) and showing the interaction. | |
|
|
Table 2 Molecular docking results of the dimetallic complexes 1a and 2b with TAR RNA (Unit: kcal/mol). |
To investigate whether the differences in the structure features in dimetallic complexes can affect RNA-binding properties, and furthermore, to gain some insight into the structure-activity relationship, in this paper, eight dimetal complexes bridged with naphthalene core have been synthesized. Two types of ligands, termed as "close" (1a-1d) and "open" (2a-2d) complexes were prepared respectively. The comparative investigations of the binding abilities towards TAR and RRE RNA were explored both theoretically and experimentally. The reactivity showed that complexes 1a and 2b can both interact with the TAR and RRE RNA in the mode electrostatic binding. However, the fluorescence experiments suggested that their abilities to bind with RNA is different. Therefore, the structure dependent binding activity was explained on the basis of the differences in the conformation. These findings should be valuable in understanding the relationship of RNA-binding behaviors of this class of bidentate complexes. Their further application as valuable lead compounds towards RNA-protein binding inhibitors is ongoing in our lab.
AcknowledgmentsThis work was supported by the National Key R&D Program of China (No. 2017YFA0208100), the National Natural Science Foundation of China (Nos. 21778057, 21502201 and 21420102003), Beijing Natural Science Foundation (No. 2162049), Young Elite Scientist Sponsorship Program by CAST (No. 2015QNRC001) and Chinese Academy of Sciences.
| [1] |
J.L. Childs-Disney, M.D. Disney, Annu. Rev. Pharmacol. 56 (2016) 123-140. DOI:10.1146/annurev-pharmtox-010715-103910 |
| [2] |
M.D. Disney, A.J. Angelbello, Acc. Chem. Res. 49 (2016) 2698-2704. DOI:10.1021/acs.accounts.6b00326 |
| [3] |
T. Hermann, WIRES RNA 7 (2016) 726-743. DOI:10.1002/wrna.1373 |
| [4] |
C.M. Connelly, M.H. Moon, J.S. Schneekloth, Cell Chem. Bio. 23 (2017) 1077-1090. |
| [5] |
S.P. Velagapudi, B.R. Vummidi, M.D. Disney, Curr. Opin. Chem. Biol. 24 (2015) 97-103. DOI:10.1016/j.cbpa.2014.10.024 |
| [6] |
M.D. Disney, G. Varani, Curr. Opin. Chem. Biol. 30 (2015) 79-88. |
| [7] |
S. Kang, K. Im, J. Baek, S. Yoon, H. Min, ChemBioChem 15 (2014) 1071-1078. DOI:10.1002/cbic.201402007 |
| [8] |
M.D. Disney, Drug Discov. Today 18 (2013) 1228-1236. DOI:10.1016/j.drudis.2013.07.024 |
| [9] |
L.R. Guan, M.D. Disney, ACS Chem. Biol. 7 (2012) 73-86. DOI:10.1021/cb200447r |
| [10] |
J. Gallego, G. Varani, Acc. Chem. Res. 34 (2001) 836-843. DOI:10.1021/ar000118k |
| [11] |
J.R. Thomas, P.J. Hergenrother, Chem. Rev. 108 (2008) 1171-1224. DOI:10.1021/cr0681546 |
| [12] |
A.C. Wolter, A.K. Weickhmann, A.H. Nasiri, et al., Angew. Chem. Int. Ed. 56 (2017) 401-404. DOI:10.1002/anie.201609184 |
| [13] |
X.H. Lv, Z.L. Ren, D.D. Li, et al., Chin. Chem. Lett. 28 (2017) 377-382. DOI:10.1016/j.cclet.2016.10.029 |
| [14] |
M.G. Costales, C.L. Haga, S.P. Velagapudi, et al., J. Am. Chem. Soc. 139 (2017) 3446-3455. DOI:10.1021/jacs.6b11273 |
| [15] |
H. Yan, U. Bhattarai, Z.F. Guo, F.S. Liang, J. Am. Chem. Soc. 139 (2017) 4987-4990. DOI:10.1021/jacs.7b00610 |
| [16] |
D. Lim, W.G. Byun, J.Y. Koo, H. Park, S.B. Park, J. Am. Chem. Soc. 138 (2016) 13630-13638. DOI:10.1021/jacs.6b06965 |
| [17] |
S.P. Velagapudi, M.D. Cameron, C.L. Haga, et al., Proc. Natl. Acad. Sci. U. S. A. 113 (2016) 5898-5903. DOI:10.1073/pnas.1523975113 |
| [18] |
Y. Katsuda, S. Sato, L. Asano, et al., J. Am. Chem. Soc. 138 (2016) 9037-9040. DOI:10.1021/jacs.6b04506 |
| [19] |
J.A. Howe, H. Wang, T.O. Fischmann, et al., Nature 526 (2015) 672-677. DOI:10.1038/nature15542 |
| [20] |
W.Y. Yang, H.D. Wilson, S.P. Velagapudi, M.D. Disney, J. Am. Chem. Soc. 137 (2015) 5336-5345. DOI:10.1021/ja507448y |
| [21] |
C.H. Wong, L. Nguyen, J. Peh, et al., J. Am. Chem. Soc. 136 (2014) 6355-6361. DOI:10.1021/ja5012146 |
| [22] |
S. Phongtongpasuk, S. Paulus, J. Schnabl, et al., Angew. Chem. Int. Ed. 52 (2013) 11513-11516. DOI:10.1002/anie.201305079 |
| [23] |
W.Y. Yang, R. Gao, M. Southern, P.S. Sarkar, M.D. Disney, Nat. Commun. 7 (2016) 11647. DOI:10.1038/ncomms11647 |
| [24] |
J.F. Arambula, S.R. Ramisetty, A.M. Baranger, S.C. Zimmerman, Proc. Natl. Acad. Sci. U. S. A. 106 (2009) 16068-16073. DOI:10.1073/pnas.0901824106 |
| [25] |
X. Li, Y. Zou, H.G. Hu, Chin. Chem. Lett. (2018). DOI:10.1016/j.cclet.2018.01.018 |
| [26] |
T.F. Fu, L. Ao, Z.C. Gao, X.L. Zhang, F. Wang, Chin. Chem. Lett. 27 (2016) 1147-1154. DOI:10.1016/j.cclet.2016.06.054 |
| [27] |
J.W. Zheng, L. Ma, Chin. Chem. Lett. 27 (2016) 627-630. DOI:10.1016/j.cclet.2016.01.052 |
| [28] |
N. Arshada, N. Abbasa, M.H. Bhatti, et al., J. Photochem. Photobiol. B 117 (2012) 228-239. DOI:10.1016/j.jphotobiol.2012.10.003 |
| [29] |
L.J.K. Boerner, J.M. Zalesk, Curr. Opin. Chem. Biol. 9 (2015) 135-144. |
| [30] |
D.P. Buck, C.B. Spillane, J.G. Collins, F.R. Keene, Mol. BioSyst. 4 (2008) 851-854. DOI:10.1039/b803216e |
| [31] |
E. Alberti, M. Zampakou, D. Donghi, J. Inorg. Biochem. 163 (2016) 278-291. DOI:10.1016/j.jinorgbio.2016.04.021 |
| [32] |
J. Malina, M.J. Hannon, V. Brabec, Sci. Rep. 6 (2016) 29674. DOI:10.1038/srep29674 |
| [33] |
B.T. Patterson, J.G. Collins, F.M. Foley, F.R. Keene, J. Chem. Soc. Dalton Trans. (2002) 4343-4350. |
| [34] |
J.A. Smith, J.G. Collins, B.T. Patterson, F.R. Keene, Dalton Trans. (2004) 1277-1283. |
| [35] |
L. Cheng, A. Mahendran, R.L. Gonzalez, R. Breslow, Proc. Natl. Acad. Sci. U. S. A. 111 (2014) 7920-7924. DOI:10.1073/pnas.1407295111 |
| [36] |
L. Cheng, K.G. Abhilash, R. Breslow, Proc. Natl. Acad. Sci. U. S. A. 109 (2012) 12884-12887. DOI:10.1073/pnas.1210846109 |
| [37] |
T.J. Thomas, H.A. Tajmir-Riahi, T. Thomas, Amino Acids 48 (2016) 2423-2431. DOI:10.1007/s00726-016-2246-8 |
| [38] |
M. Menzi, H.L. Lightfoot, J. Hall, Future Med. Chem. 7 (2015) 1733-1749. DOI:10.4155/fmc.15.90 |
| [39] |
K.A. Howard, J. Kjems, Opin Expert, Biol. Ther. 7 (2007) 1811-1822. |
| [40] |
T. Jany, A. Moreth, C. Gruschka, et al., Inorg. Chem. 54 (2015) 2679-2690. DOI:10.1021/ic5028465 |
| [41] |
S.N. Richter, G. Palu, Curr. Med. Chem. 13 (2006) 1305-1315. DOI:10.2174/092986706776872989 |
| [42] |
M. Yang, Infect. Disord. Drug Targets 5 (2005) 433-444. DOI:10.2174/156800505774912901 |
| [43] |
S. Bannwarth, A. Gatignol, Curr. HIV Res. 3 (2005) 61-71. DOI:10.2174/1570162052772924 |
| [44] |
C. Jain, J.G. Belasco, Mol. Cell 7 (2001) 603-614. DOI:10.1016/S1097-2765(01)00207-6 |
| [45] |
M.D. Daugherty, D.S. Booth, B. Jayaraman, Y. Cheng, A.D. Frankelc, Proc. Natl. Acad. Sci. U. S. A. 107 (2010) 12481-12486. DOI:10.1073/pnas.1007022107 |
| [46] |
L. Qi, J.R. Wei, X.J. Lv, Y. Huo, Z.Q. Zhang, Biosens. Bioelectron. 86 (2016) 287-292. DOI:10.1016/j.bios.2016.06.051 |
| [47] |
F.A. Abulwerdi, M.D. Shortridge, J. Sztuba-Solinska, et al., J. Med. Chem. 59 (2016) 11148-11160. DOI:10.1021/acs.jmedchem.6b01450 |
| [48] |
J. Sztuba-Solinska, S.R. Shenoy, P. Gareiss, et al., J. Am. Chem. Soc. 136 (2014) 8402-8410. DOI:10.1021/ja502754f |
| [49] |
G.M. Morris, R. Huey, W. Lindstrom, et al., J. Comput. Chem. 30 (2009) 2785-2791. DOI:10.1002/jcc.v30:16 |
| [50] |
A. Robertazzi, A.V. Vargiu, A. Magistrato, et al., J. Phys. Chem. B 113 (2009) 10881-10890. DOI:10.1021/jp901210g |
| [51] |
K. Zheng, F. Liu, X.M. Xu, et al., New J. Chem. 38 (2014) 2964-2978. DOI:10.1039/C4NJ00092G |
| [52] |
Y. Yoshikawa, K. Kobayashi, S. Oishi, et al., Bioorg. Med. Chem. Lett. 22 (2012) 2146-2150. DOI:10.1016/j.bmcl.2012.01.134 |
| [53] |
A. Kumar, K.Y. Zhang, J. Chem. Inf. Model. 53 (2013) 1880-1892. DOI:10.1021/ci400052w |
| [54] |
M.L. Verdonk, G. Chessari, J.C. Cole, et al., J. Med. Chem. 48 (2005) 6504-6515. DOI:10.1021/jm050543p |
 2018, Vol. 29
2018, Vol. 29